Building and Analyzing CardioKG: A Comprehensive Knowledge Graph of Cardiovascular Drugs, Diseases, Proteins, and Pathways.
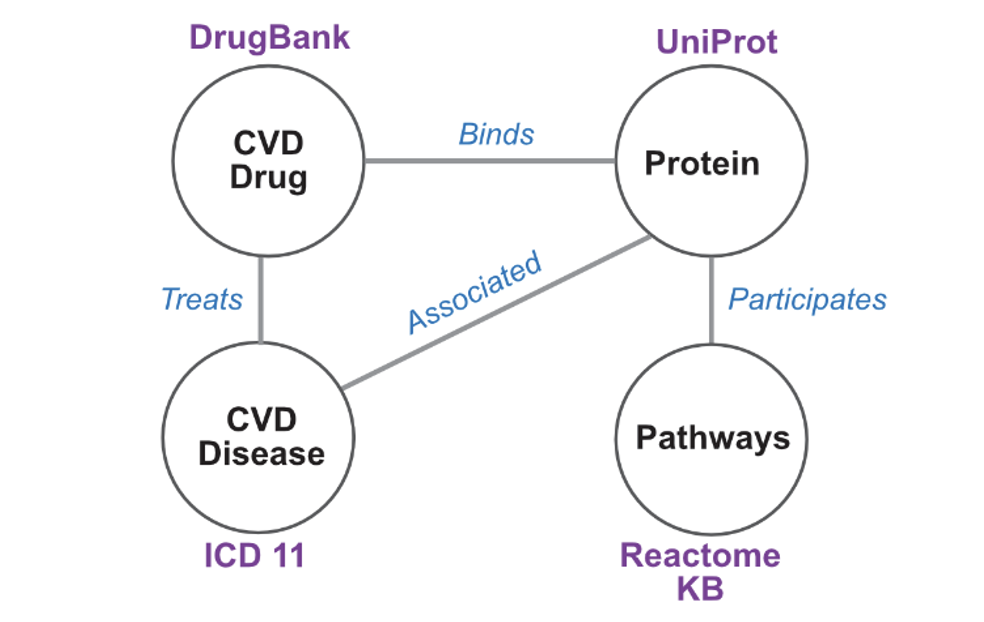
This project includes the development of a Graph Neural Network (GNN) model with a knowledge graph in the interface of Cardiovascular Diseases (CVD) and CVD drugs,drug target proteins with underlying molecular mechanism (e.g., pathways).The knowledge graph will be utilized for graph embedding so that it can be used for the Graph Neural Network model. In general, GNN offers three different prediction models: (1) node prediction, (2) link prediction, and (3) graph classification. We are particularly interested in building the model for 'link prediction' between CVD drugs and proteins and pathways terms.
- Novel drug target prediction: Using the knowledge graph to explore the relationship between a cardiovascular disease (CVD) and drugs linked to protein drug targets, which may share multiple biological pathways with many proteins, to help identify potential novel drug targets.
- Drug repurposing: Find potential new uses of existing drugs by linking drugs with other disease phenotypes that are not currently designated for the drug by examining the complex subgraph of a graph network.
Heterograph convolution is designed by combining the individual relationship model.
conv = HeteroConv({
('drug', 'associates', 'disease'): GCNConv(-1, hidden_channels),
('disease', 'assigns', 'pathways'): SAGEConv((-1, -1), hidden_channels),
('protein', 'candidate', 'pathways'): GATConv((-1, -1), hidden_channels),
('protein', 'associated', 'drug'): GATConv((-1, -1), hidden_channels),
}, aggr='sum')
- Understand the schema, data content, and development of the knowledge graph in the interface of Cardiovascular Disease, CVD drus, drug target proteins and underlying molecular mechanism.
- Learn more about graph embedding functionality in Neo4j GDS library and DGL-KE library.
- Learn more about the fundamentals of machine learning models ( e.g., allocating data for training, validation, and test, selecting proper GNN message passing algorithm, selecting optimizer, Cost function, accuracy metric, and inferences).
- Explore about homogeneous Graph Neural Network with provided tutorial-1.
- Explore heterogeneous Graph Neural Networks with provided tutorial-2.
- Develop the graph embedding for the heterogeneous knowledge graph.
- Prepare the training, validation, and testing by masking the nodes or edges.
- Train the model with tuning hyperparameters and interpret the performance.
- Implement the model for link prediction and analyze it with biomedical findings.
This project offers the wonderful opportunity to learn and build cutting-edge machine learning models in graph data. Participants will get hands-on experience with GNN and KG libraries (e.g., Neo4j, DGL, and Pytorch Geometric).
Understanding the role of drug in Cardiovascular Disease through a molecular mechanism and adverse effects associated drug target proteins is the core of the scientific goal of the project. Prediction of the possible links between Drug and Drug target proteins/ Drug and Diseases opens a new research horizon. Newly predicted associations could provide further insight into the molecular mechanism and disease pathogenesis.