Cardiovascular disease is one of the most important diseases in the world. People are trying to explore it deeper and deeper for a long time. With the development of big data explosion and artificial Intelligence method, now we have more strategies to help us explore the disease. For medicine data, more and more graph data is produced, which is better to draw the relationship map between entities. The aim of this project is to explore the cardiovascular diseases related molecular mechanisms following MeSH descriptors using the graph neural network (GNN).
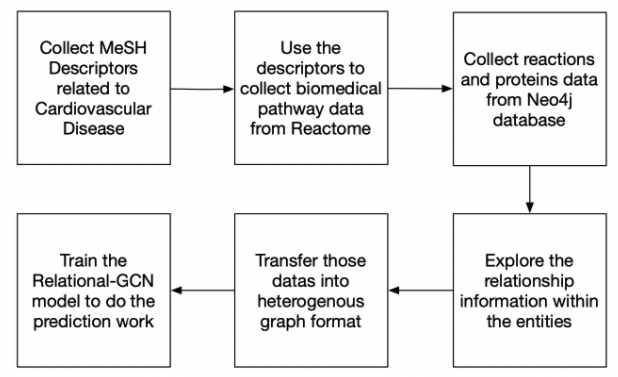
Until now, there are no open source pathway data related to cardiovascular diseases. As a result, we collected them by ourselves. Reactome database is a pathway database which provides intuitive bioinformatics tools for research. As a result, we used the API on the website and applied web crawler strategy to collect the data we needed. Also, we use the Neo4j database, which is a graph database management system, to transfer the data format into what we want. Then we utilize the Cypher query to explore the relationship inside the data we collected and finally set the dataset successfully.
This is an GCN(Graph Convoluted Network) project to classify every pathway related to cardiovascular diseases into different sub-class diseases. Because entities, such as pathway, reaction, protein and so on, inside our body are connected with each other tightly, we could build a graph dataset to store them and use the information inside the graph to give the right class for every pathway. It is a supervised learning model. We need class labels for part of the pathways and use them and the graph to predict the rest of the pathways. This project will be helpful to precision medicine. When a pathway damage is detected, we can know what kind of precise diseases the patient has.
We have two prediction model. Model 1 can predict the high level sub-class diseases for each pathway belongs to. Model 2 can predict the most possible precise diseases for each pathway belongs to. This result can help us identify what kind of disease the exact pathway will affect. The input of our models are the relational graph we build. The output of our models will be the class for each pathway without label. The class information is expected to be recovered from the neighborhood structure of the graph. It’s extended with multi-edge encoding to compute embedding of the entities.
Requires DGL. This can be installed with pip as : pip install DGL
1. Collect all the MeSH Descriptors related to Cardiovascular Disease(CVD) from MeSH Tree and load the graph data from Reactome website. The CVD tree data can be found /neo4j/cvd.csv. The Reactome graph data can be download from here. The codes to extract CVD form MeSH tree can be found at /neo4j/CVD-Tree.ipynb. The codes to load Reactome graph data on local Neo4j can be found at /neo4j/CVDTree+Reactome.ipynb.
2. Use those MeSH Descriptors to do the web crawler by advanced search from Reactome website to collect all the pathway data belong to each MeSH Descriptors(precise diseases). The final pathway data related to cardiovascular diseases can be found /AdvancedSearch/pathwayID_MeSH.csv. The statistics results about the result can be found at /AdvancedSearch/Count_pathway.csv and /AdvancedSearch/pathwayStat.csv. The web crawler codes can be found at /AdvancedSearch/advancedSearch.ipynb.
3. Apply Cypher query to collect reactions and proteins data related to each pathway on Neo4j graph database. And also extract the preceding event reactions information. The relationship information between pathway and reaction, reaction and reaction, pathway and MeSH Descriptor can be found at /relationshipExplore/path_reac.h5, /relationshipExplore/reac_reac.h5 and /relationshipExplore/mesh_path.h5. The codes to extract the relationship information can be found at '/relationshipExplore/relationshipExplore.ipynb`.
1. Use DGL library to construct the heterogeneous graph on our graph data due to the various edge and node types. The information about the graph should be showed as below:
Node types: ['meshDescriptor', 'pathway', 'reaction', 'superclass']
Edge types: ['conclude', 'hasEvent', 'precedingEvent', 'belongto', 'belong', 'eventOf', 'laterEvent', 'include']
Canonical edge types: [('meshDescriptor', 'conclude', 'pathway'), ('pathway', 'hasEvent', 'reaction'), ('reaction', 'precedingEvent', 'reaction'), ('meshDescriptor', 'belongto', 'superclass'), ('pathway', 'belong', 'meshDescriptor'), ('reaction', 'eventOf', 'pathway'), ('reaction', 'laterEvent', 'reaction'), ('superclass', 'include', 'meshDescriptor')]
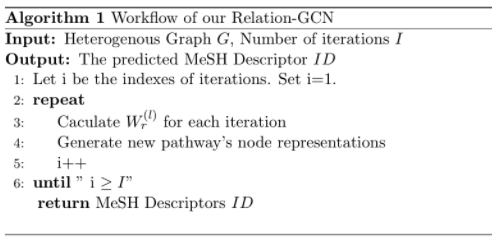
2. Use relational Graph Convolutional Networks which information passing functions concludes both edge and node information.
3. Build two models, one that can predict the high level MeSH descriptor class for each pathway belong to and the other that can predict the most possible MeSH descriptors for the pathway belong to.
4. Cut our dataset into 8:1:1 as train dataset, validation dataset and test dataset.
5. For our prediction result, we achieve 92% and 75% accuracy on our test dataset for model 1 and model 2 respectively.
The input of the model is the graph parameters. The output of the model is the vector of class label for test dataset pathway.
For example, for model 2, there are ten pathways in the dataset. So the output vector is (0,0,2,2,3,1,0,0,0,1).
0:Cardiovascular Abnormalities
1:Cardiovascular Infections
2:Heart Diseases
3:Vascular Diseases
All the related codes can be found at /DGL/GraphBuild_model1.ipynb and /DGL/GraphBuild_model2.ipynb
Created by Wangjie Zheng while working with Dibakar Sigdel in the Ping Lab at UCLA, Summer 2020