Nextflow pipeline to aggregate pertinent metrics across pipeline runs on the Seqera Platform.
The pipeline performs the following steps:
- Downloads run information via the Seqera CLI in parallel
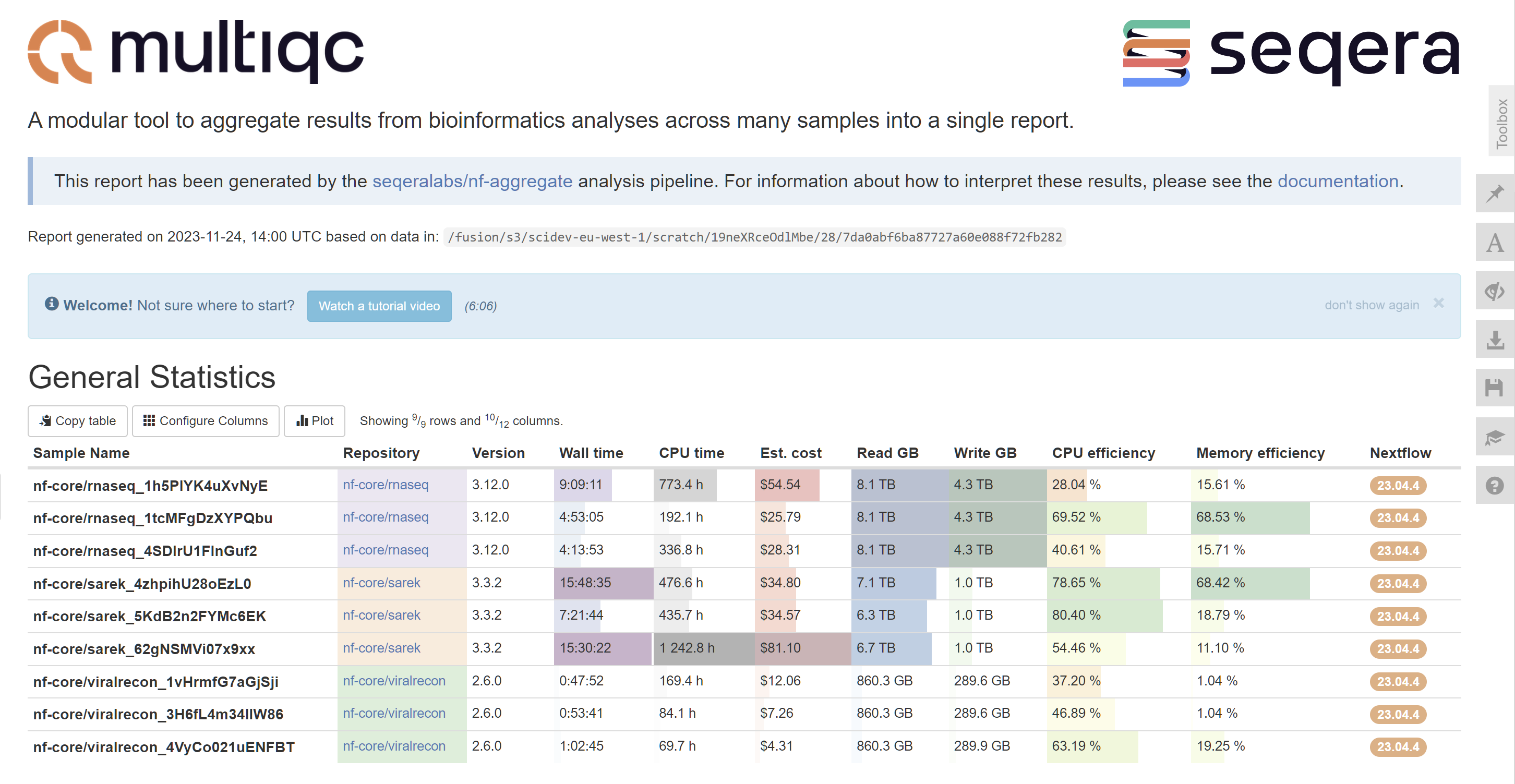
- Runs MultiQC to aggregate all of the run metrics into a single report
You can download an example MultiQC report here.
The primary input to the pipeline is a file containing a list of run identifiers from the Seqera Platform. These can be obtained from details in the runs page for any pipeline execution. For example, we can create a file called run_ids.csv with the following contents:
id,workspace
4Bi5xBK6E2Nbhj,community/showcase
4LWT4uaXDaGcDY,community/showcase
38QXz4OfQDpwOV,community/showcase
2lXd1j7OwZVfxh,community/showcase
This pipeline can then be executed with the following command:
nextflow run seqeralabs/nf-aggregate \
--input run_ids.csv \
--outdir ./results \
-profile docker \
The results from the pipeline will be published in the path specified by the --outdir and will consist of the following contents:
.
├── gantt/
│ ├── nf-core_rnaseq/
│ │ └── 4LWT4uaXDaGcDY_gantt.html ## Gantt plot for run
├── multiqc/
│ ├── multiqc_data/
│ ├── multiqc_plots/
│ └── multiqc_report.html ## MultiQC report
├── pipeline_info/
└── runs_dump/
├── 4Bi5xBK6E2Nbhj/ ## Output of 'tw runs dump'
├── service-info.json
├── workflow-launch.json
├── workflow-load.json
├── workflow-metrics.json
├── workflow-tasks.json
└── workflow.json
Please feel free to create issues and pull requests to this repo if you encounter any problems or would like to suggest/contribute feature enhancements.
nf-aggregate was written by the Scientific Development and MultiQC teams at Seqera Labs.
The nf-core pipeline template was used to create the skeleton of this pipeline but there are no plans to contribute it to nf-core at this point.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.