This repository contains an implementation of the Whole Exome Sequencing (WES) pipeline based on GATK best practices workflows using WDL scripts (Workflow Description Language).
- Optimized to run samples in parallel
- The Docker version allow users to chose the number of samples to run in parallel based on available resources (threads and memory; available upon request)
- WDL and JSON made easy by removing "unecessary statements"
- Single line command to run the whole pipeline (QC, trimming, mapping, markduplicates, base recalibration, variant calling, annotation)
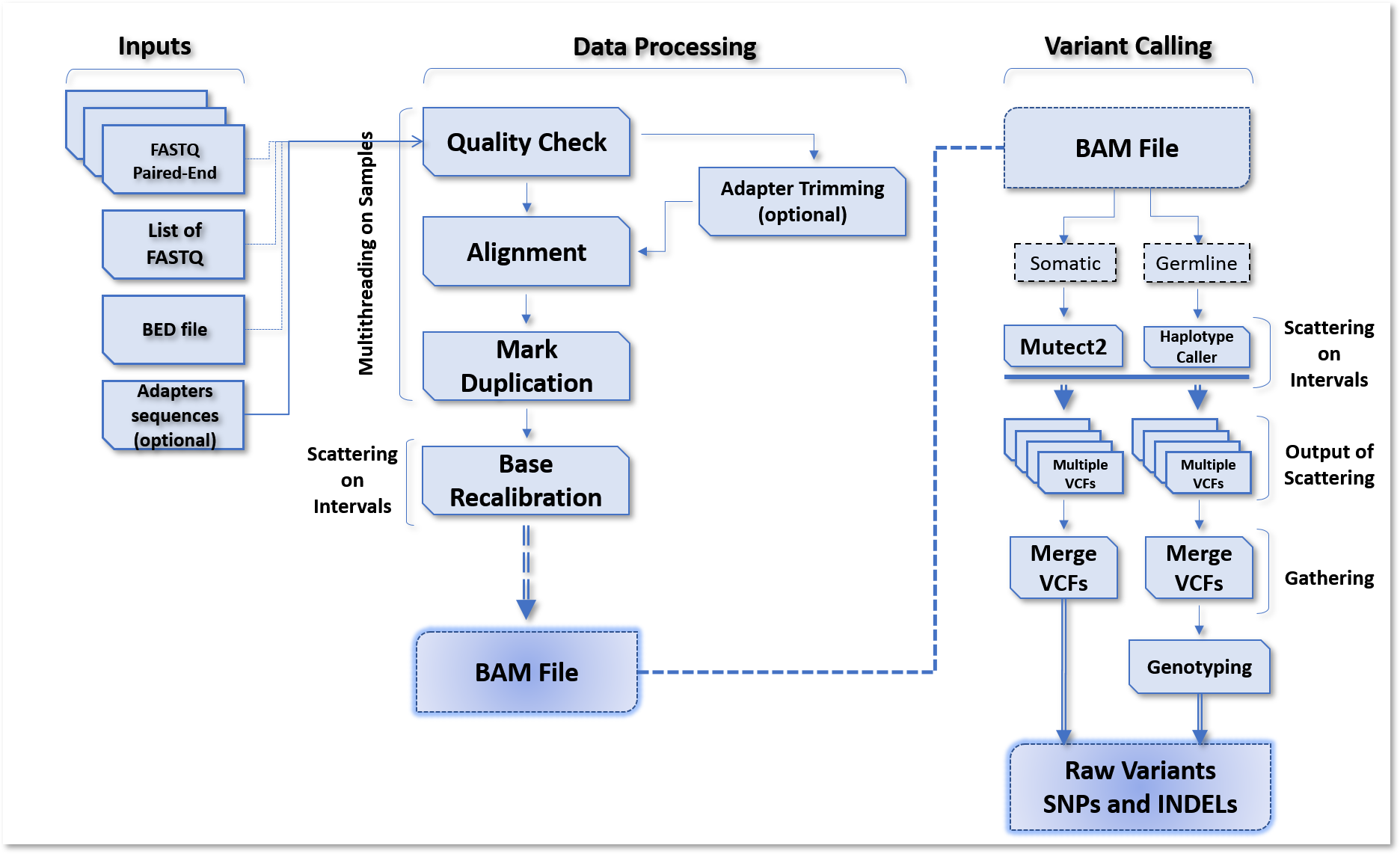
The diagram below summarizes the germline and somatic analysis (tumor only or tumor/normal).
The pipelines consist of WDL scripts that run the analysis in addition to shell scripts that act as intermediate steps. The pipelines were tested successfully based on the following dependencies:
Java 8
Cromwell v36
FastQC v0.11.5
BWA 0.7.17-r1194-dirty
Cutadapt 1.18
Samtools 1.8 – should be installed in the PATH
GATK-4.0.11.0
Tabix 0.2.5
Also, you should download the human reference genome and index it using BWA. In addition, some databases should be downloaded too:
dbsnp
phase1snps
Mills_and_1000G_gold_standard
HapMap
Omni
Axiom
You can download the reference genome and its index, the intervals and the databases listed above from resources directory provided by Broad Institute from the following link:
https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0/?pli=1
Each one of the WDL and shell scripts can be invoked independently by providing the project directory as argument.
the projectDir should have the following structure:
1- A directory named "fastq" which contains FASTQ files. FASTQ files should have the following naming style:
sampleName_R1.fastq.gz and sampleName_R2.fastq.gz
2- A directory named "lists" containing three files:
1) fastq_list.txt: A tab separated file listing samples in the following format:
sampleName1 sampleName1_R1.fastq.gz sampleName1_R2.fastq.gz
sampleName2 sampleName2_R1.fastq.gz sampleName2_R2.fastq.gz
2) intervals.txt Contains a list of full path of all intervals in BED format:
path/to/intervals/scattered_calling_intervals/temp_0001_of_50/scattered.interval_list
path/to/intervals/scattered_calling_intervals/temp_0002_of_50/scattered.interval_list
path/to/intervals/scattered_calling_intervals/temp_0003_of_50/scattered.interval_list
path/to/intervals/scattered_calling_intervals/temp_0004_of_50/scattered.interval_list
path/to/intervals/scattered_calling_intervals/temp_0005_of_50/scattered.interval_list
3) adapters.txt Contains adapters to be trimmed:
The first line should contain first read adapter (forward) and the second
line should contain second read adapter (reverse):
CTGTCTCTTGATCACA
TGTGATCAAGAGACAG
To run the pipeline, you must specify full paths for each tool and database in the JSON file. Once done, you can invoke the pipeline using the following command:
/path/to/run.sh /path/to/project/directory /path/to/cromwell.jar
To use the Docker image (available upon request), you must prepare the ‘project directory’ as mentioned above and invoke the Docker image using the following command:
docker run -it -v /path/to/project/directory/:/data/ pklab/wes_pipelines
We can invoke each WDL and shell scripts separately.
If we use the Docker, all you need is to use fastq_list.txt, intervals.txt and adapters.txt from the lists directory.