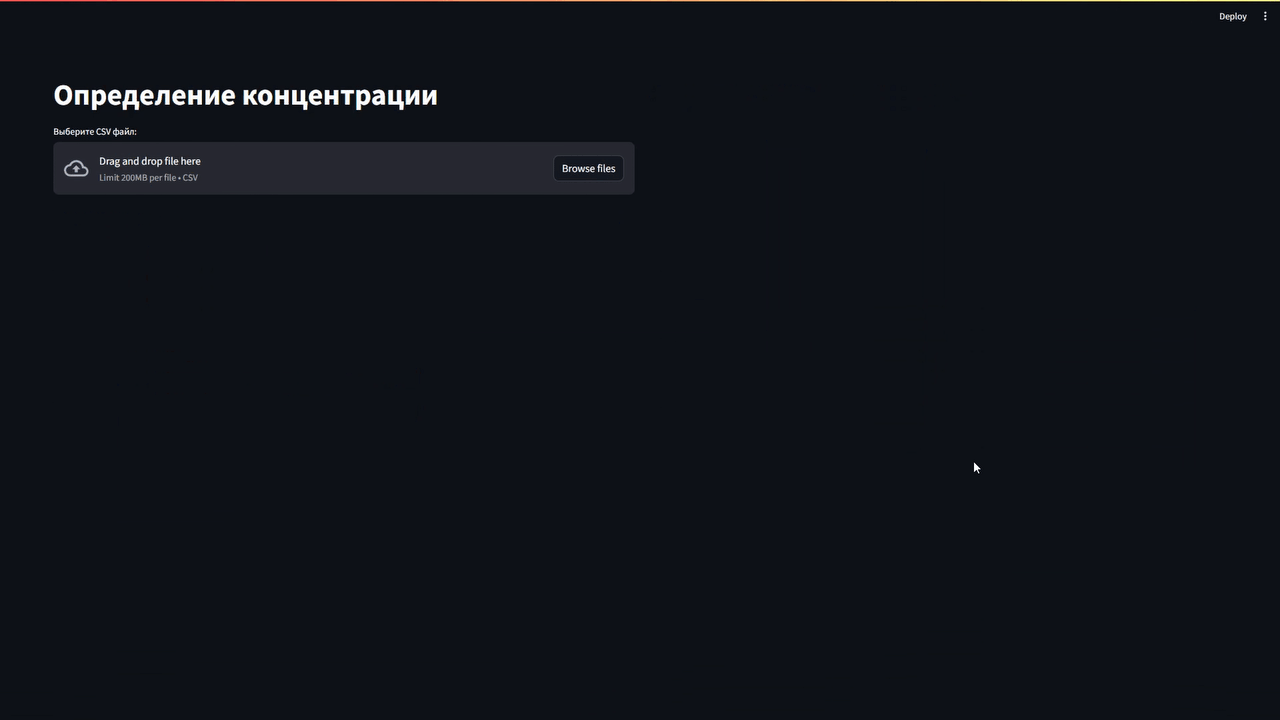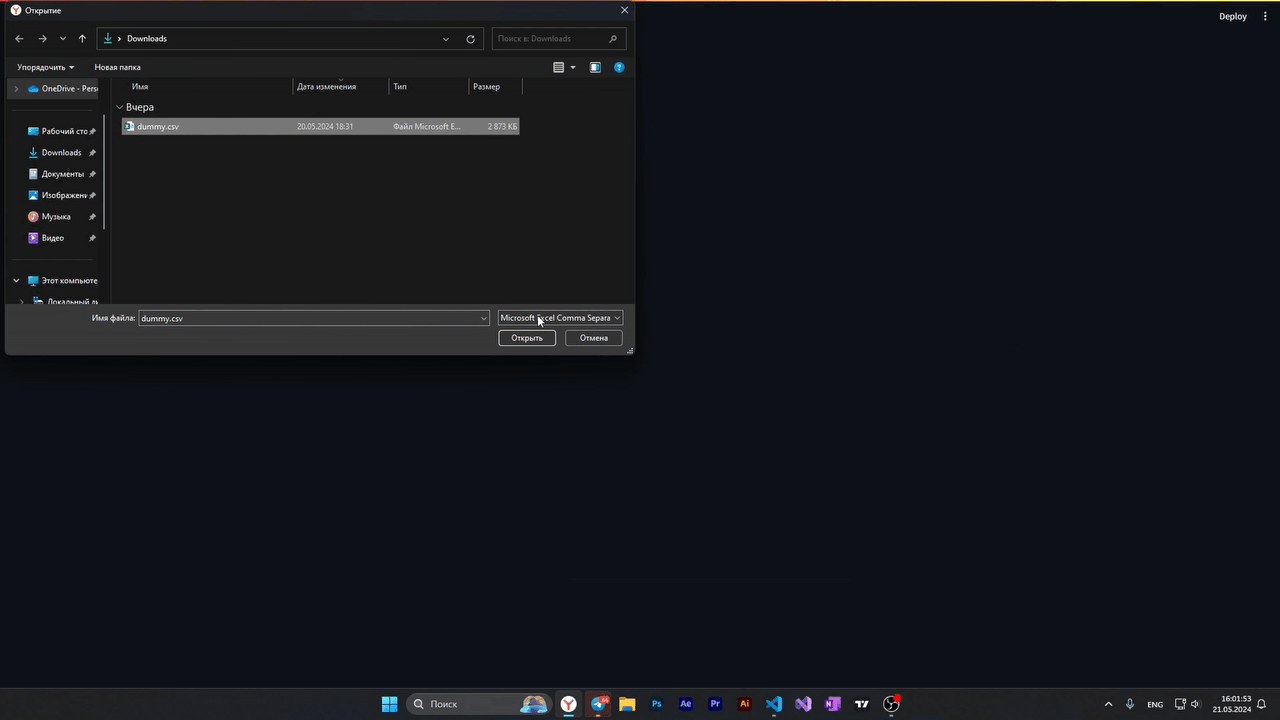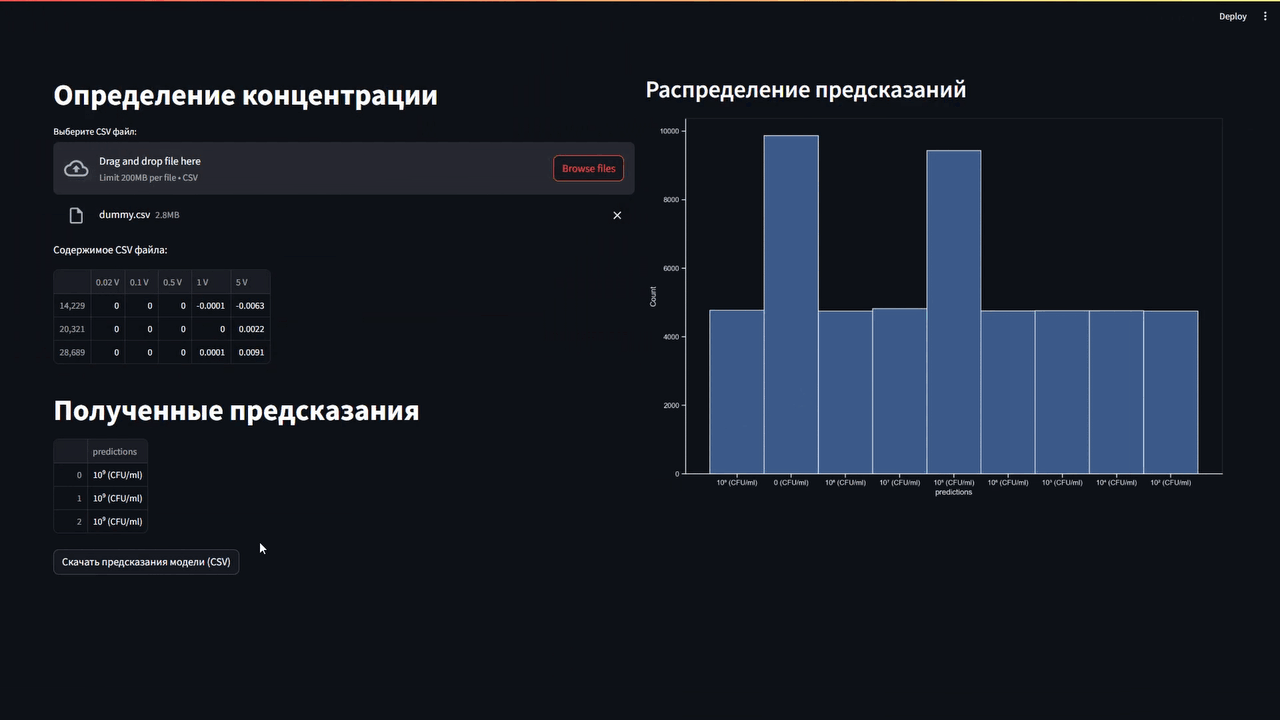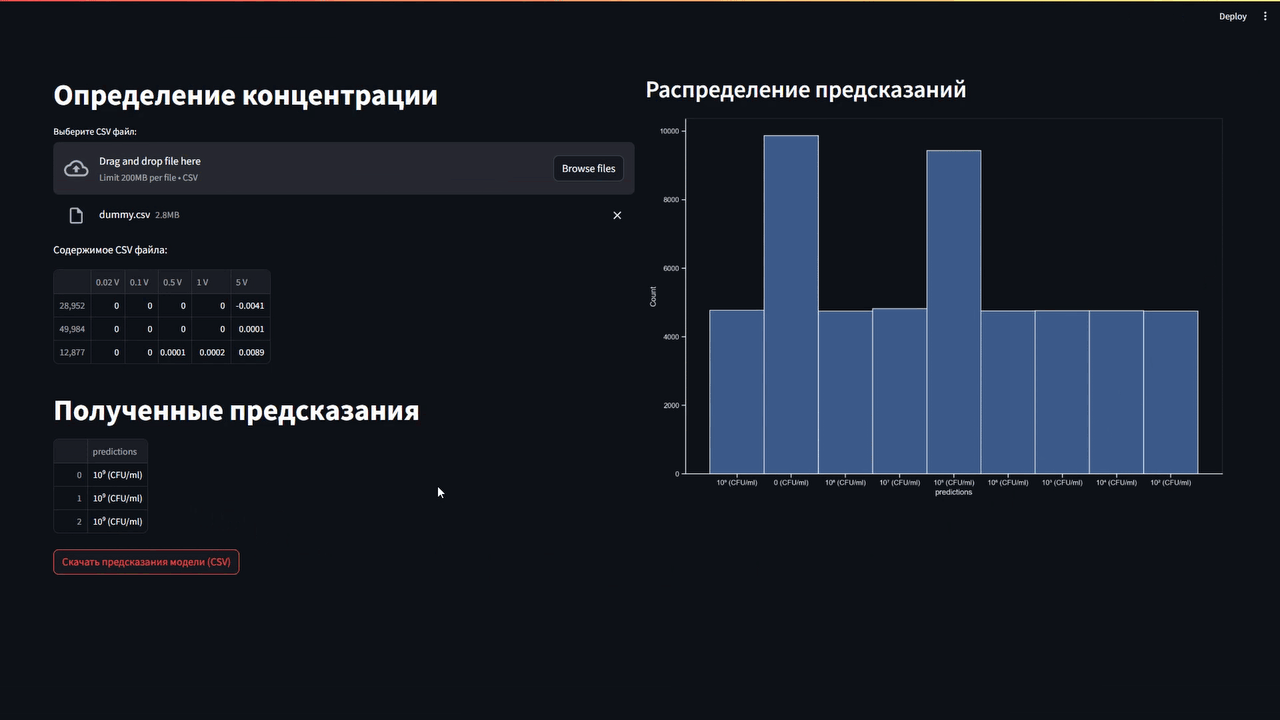
Версия README.md на русском: В репозитории
Author: Zhamkov N. D.
Tutor: Dyakonov А. V.
With the recent commit you are able to run inference using local web-app on streamlit. For now, web-app works only on compound inference task for e-coli bacteria. Soon to be upgraded.
- Clone this repo using:
git clone https://github.com/plugg1N/itmo-ml-research.git- Install requirements for this project:
pip3 install -r requirements.txt- Run streamlit app in
srcfolder:
streamlit run web.py If first didn't work:
python3 -m streamlit run web.pyIn the modern world, the problem of rapid and accurate determination of the concentration and type of various bacteria is significant in the fields of medicine and food industry. Currently, for these purposes, the classical Koch method is often used, which, although standard in the field, is criticized for its low detection rate (24-48 hours). Therefore, the development of more efficient detection methods is highly relevant. In our research work, we propose an innovative method aimed at determining the concentration and type of bacteria using advanced machine learning techniques. The proposed method can not only solve this problem but also contribute significantly to improve the detection and classification processes of bacteria. In this paper, existing detection techniques are analyzed and their shortcomings are identified. The advantages of a new approach based on the use of machine learning technologies are described, providing high accuracy of results and a significant reduction of the process, from 24-48 hours to ~1 hour. The aim of the research is to develop and implement a method for bacterial concentration determination and classification using machine learning technologies. This project analyzes different machine learning models and identifies the model with the best detection results.
In this work:
- Existing methods for determining the concentration and type of bacteria were analyzed.
- The VAC characteristics of media with different bacterial strains were collected and databases were compiled.
- Optimal machine learning models for bacterial concentration determination and classification are identified. The parameters of machine learning algorithms were selected.
- The evaluation metrics of the models were compared.
We proved the hypothesis that using a machine learning model as a way to determine the concentration of bacteria and their types significantly speeds up the detection process while maintaining accuracy comparable to generally accepted standards (Koch's method). The model we have trained can be easily integrated into food industry production processes, allowing the detection of microbial concentrations in raw materials and products with high speed and accuracy. The detection process will take less than an hour, with >98% accuracy in determining the concentration and type of bacteria, resulting in a significant reduction in the risk of disease and complications caused by pathogens.
Koch's method involves serial dilutions of the test material in molten agar followed by transfer of the agar with the diluted culture to a Petri dish.
However, despite its wide applicability and accuracy, the classical Koch method has the significant disadvantage of a long turnaround time. In settings where rapid and accurate analysis of pathogen concentrations is required, an approach that requires from 24 up to 48 hours and moremay be inefficient and impractical.
- Accuracy
- Precision
- Recall
- F1-score
We're more interested in Recall, since we're betting on making fewer mistakes for a positive (possibly dangerous) class rather than just having high accuracy in general.
Volt-ampere Characteristics (VAC) represent the relationship between current and voltage in an electrical system. In the context of bacterial research, VAC is used, for example, in electrochemical impedance spectroscopy (EIS). for example, in electrochemical impedance spectroscopy (EIS). This technique allows measure changes in electrical impedance in a system, including a biological medium with microorganisms, under the influence of an alternating electric field. Given the behavior of bacteria, the use of cyclic voltammetry to detection and enumeration of bacteria can be effective due to its advantages: fast response and simple equipment. A conductometric method such as EIS does not require tags as a means of rapid detection of bacteria, which simplifies sensor preparation. There are systems based on hydrogel-based systems used to detect a variety of biological molecules. This technique is already well established as a selective method for analyzing tick-borne virus encephalitis virus. A flexible hydrogel/liquid metal detector is used to monitor bacteria in various media and products. Such a device provides faster and cheaper quantitative analysis of bacteria than existing methods.
Prior to the experiment, we grew the bacteria at 37°C in broth LB for 24 hours. Then, in order to determine the number of live bacteria or colony forming units (CFU), we prepared a series of serial tenfold dilutions. The dilutions were achieved by serial dilution of the bacterial inoculum in 10 tubes, each containing 9 mL of physiologic solution (dilutions ranging from 10-1 to 10-9) . All dilutions were then seeded 1 ml at a time onto previously prepared sterile plates with LB nutrient medium, using the pouring method, and incubated at 37°C for 48 hours. incubated at 37°C for 48 hours.
Data was collected using a Keithley 6430 potentiostat. The training data set was structured with 40 separate samples for each class. Five cyclic voltammetry analysis (CVAs) were recorded for each sample. Each set of electrochemical responses was characterized by a series of 400 current values (200 forward, 200 reverse points). The test data set consisted of three samples for each class. The following potential ranges were applied in the voltage-variation mode:
- from -0.02 to 0.02 V
- from -0.1 to 0.1 v
- from -0.5 to 0.5 v
- from -1 to 1 V
- from -5 to 5 V
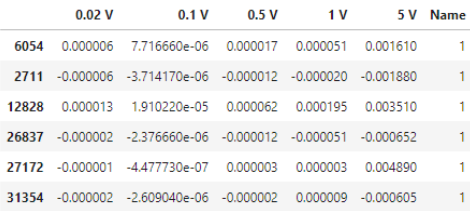
The obtained VAC data were collected and provided as a table in *.csv format (comma-separated values). The information was processed using Pandas data analysis library. Python 3.10.6 was chosen as the programming language.The data was divided into two databases. One database stored information about the VAC E. coli, The second stored information on fermented milk bacteria (B.coagulans and S. thermophilus).
We decided to apply "UnderSampling" in order to prevent class imbalance (a phenomenon in which the number of objects of one class is greater than the number of objects of of other classes). In other words, we have removed the imbalance by reducing the number of objects of the of the superior type to the smallest value by leveling the number of objects in all the data:
Next, "scalers" and "normalizers" were applied to the data to bring the factor values to a certain range. "Scalers" help machine learning algorithms machine learning algorithms such as SVM (Support Vector Machines) and K-NN (K-Nearest Neighbors), since since these algorithms are based on the distance between data points. Also, scaling 16 helps with stabilizing the sensitivity of the machine learning model to initial values. Additionally, a single dataset was created that only holds information about the VACs and the name of the bacterium itself. This dataset will be responsible for testing the hypothesis that the training the algorithm on the BAC is able to classify the type of bacterium.
The most accurate models, according to microbiology research papers, are SVM, RFC (Random Forest Classifier) and ANN (Artificial Neural Network). Since these models have performed the best, they will be among the top algorithms to validate. Also on the list of models for accuracy assessment will be "bousting" models.
Expanding models list:
- Random Forest Classifier (RFC)
- Support Vector Classifier (SVC)
- TabNet (TN)
- Extra Trees Classifier (ETC)
- Catboost Classifier (CBC)
- Logistic Regression (LR)
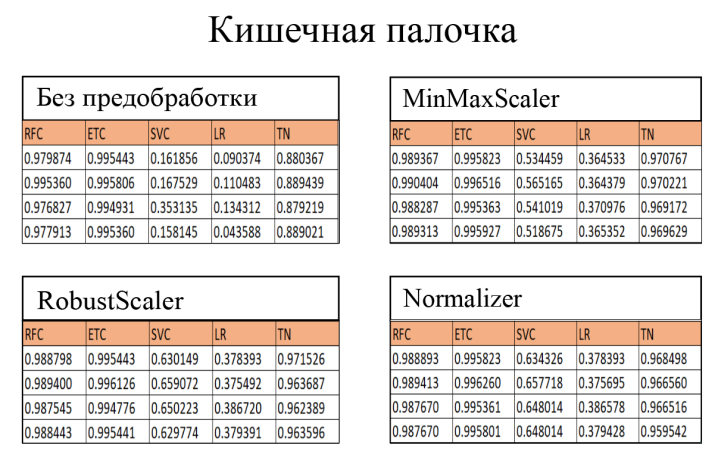
Note that of the four preprocessing methods, RobustScaler and MinMaxScaler perform best. It's complicated to determine the best preprocessing method since all the methods work differently.
From the results, we conclude that SVC and LR are not competitive models for bacterial concentration detection using VAC, as the accuracies of these models are noticeably lower than the other evaluated algorithms. The TabNet deep learning model trained on 600 epochs produces a Recall metric result no higher than 0.9849, which is not the optimal result for bacterial concentration detection.
RFC and ETC after cross-val:
- Random Forest Classifier: 0.991555
- Extra Trees Classifier: 0.997550
You may notice that "CatBoost Classifier" is not listed in the results. This bousting model was optimized and its parameters were picked up using GridSearchCV. Parameters for the search: depth, od_wait, l2_leaf_reg, iterations, leanring_rate. Results:
Of the top three models, the Catboost Classifier> (CBC) is the most optimal because this model has a large range of parameters to fit; is least susceptible to overtraining; and is able to perform the training process using the GPU and, thereby speeding up the training and/or fitting process.
The list of models and preprocessing methods remain the same, except for metrics. The average of the metrics will be averaged over the two classes, since a binary classification task is performed.The target variable becomes the "Name" column instead of "Compound". 1 - Escherichia coli, 0 - Sour milk bacteria. The new dataset looks as follows:
The LR and SVC models have, on average, higher accuracy in bacterial type classification, than in bacterial concentration detection, but these algorithms are still not competitive with other models. TN has a reasonably high accuracy, but this model is not the most optimal in the context of bacterial type determination.
RFC and ETC remain the leaders in terms of accuracy. Let's check their accuracy on cross-validation:
- Random Forest Classifier: 0.99549651
- Extra Trees Classifier: 0.99797328
We make an unambiguous conclusion that the application of the model "Catboost Classifier" as an algorithm for determining the concentration of microorganism and its type is the most optimal. optimal. This model, according to the results of the experiment, proved to be one of the most accurate models tested. It's worth mentioning that Catboost is prone to overtraining much less than RFC or ETC. High accuracy of the retrained model of the test sample on one dataset may not always mean high accuracy on another dataset..
The use of machine learning models in determining bacterial concentrations and classifying them proved to be significantly more effective than traditional methods such as the classical Koch method. In addition, "CatBoost Classifier" proved to be the most optimal model for the classification task, outperforming other algorithms not only in terms of accuracy metrics - with Recall equal to ~0.98641 - but also in terms of the number of parameters to be optimized. The most optimal preprocessing methods were "RobustScaler" and "MinMaxScaler", improving the accuracy of the algorithms by about 0.5% on average. The experiments confirmed the high efficiency of applying machine learning algorithms in practice, demonstrating high speed and accuracy of classification. The hypothesis about the effectiveness of machine learning models in detecting the concentration of microorganisms and their type was successfully confirmed.
The prospect of the project is to further investigate and improvement of methods for determining the concentration and classification of bacterial types. Key areas of focus include validation and optimization of machine learning models to improve accuracy and efficiency. In addition, expanding the scope of the study on alternative biomaterials will allow us to evaluate the performance of our algorithm on unstudied microorganisms. An important milestone is the ability to integrate the developed system into industrial processes, which opens perspectives for use in real-life conditions of production. Thus, the project not only provides innovative methods of detection, but also but also has the potential for practical application.