- Introduction
- Installation
- Quick start
- Workflow
- Reference
Did you know that Terpene Synthases (TPSs) are responsible for the most natural scents humans have ever experienced [1]? Among other invaluable molecules, TPSs are also responsible for the Nobel-prize-winning antimalarial treatment artemisinin [2] with a market size projected to reach USD 697.9 million by 2025 [3], or TPSs are accountable for the first-line anticancer medicine taxol with billion-dollar pick annual sales [4].
Welcome to the GitHub repository showcasing state-of-the-art computational methods for Terpene Synthase (TPS) discovery and characterization.
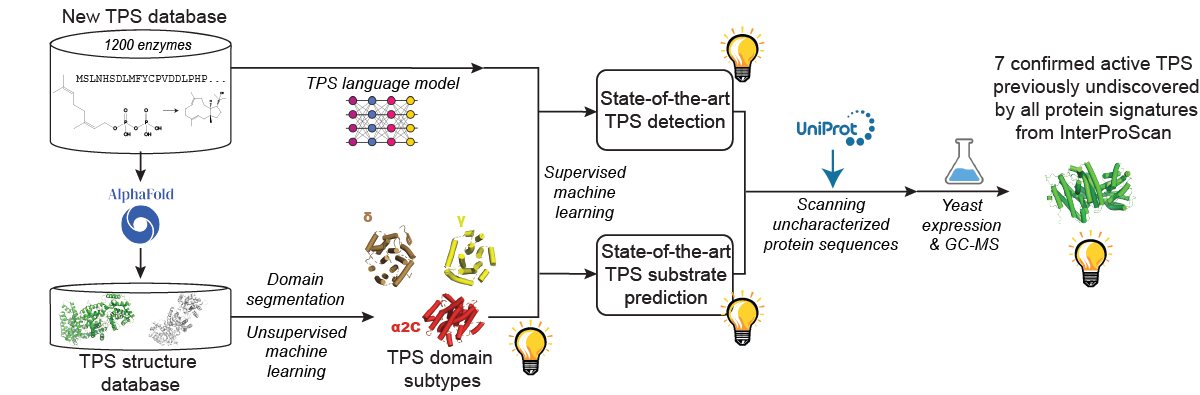
TPSs generate the scaffolds of the largest class of natural products (more than 96.000 compounds), including several first-line medicines [5]. Our research, outlined in the accompanying paper Highly accurate discovery of terpene synthases powered by machine learning reveals functional terpene cyclization in Archaea, addresses the challenge of accurately detecting TPS activity in sequence databases.
Our approach significantly outperforms existing methods for TPS detection and substrate prediction. Using it, we identified and experimentally confirmed the activity of seven previously unknown TPS enzymes undetected by all state-of-the-art protein signatures integrated into InterProScan.
Notably, our method is the first to reveal functional terpene cyclization in the Archaea, one of the major domains of life [6]. Before our work, it was believed that Archaea can form prenyl monomers but cannot perform terpene cyclization [7]. Thanks to the cyclization, terpenoids are the largest and most diverse class of natural products. Our predictive pipeline sheds light on the ancient history of TPS biosynthesis, which "is deeply intertwined with the establishment of biochemistry in its present form" [7].
Furthermore, the presented research unveiled a new TPS structural domain and identified distinct subtypes of known domains, enhancing our understanding of TPS diversity and function.
This repository provides access to our approach's source codes. We invite researchers to explore, contribute, and apply our approach to other enzyme families, accelerating biological discoveries.
git clone https://github.com/pluskal-lab/TerpeneMiner.git
cd TerpeneMiner
. scripts/setup_env.sh
conda activate terpene_miner
pip install .cd TerpeneMiner
conda activate terpene_miner
python scripts/easy_predict_sequence_only.py --input-fasta-path data/af_inputs_test.fasta --output-csv-path test_seqs_pred.csv --detection-threshold 0.2 --detect-precursor-synthaseWe sample negative (non-TPS) sequences from Swiss-Prot, the
expertly curated UniProtKB component produced by the UniProt consortium.
For reproducibility, we share the sampled sequences in data/sampled_id_2_seq.pkl.
If you want to sample Swiss-Prot entries on your own, download Swiss-Prot .fasta file
from UniProt.org Downloads to the data folder and then run
cd TerpeneMiner
conda activate terpene_miner
mkdir -p outputs/logs
if [ ! -f data/sampled_id_2_seq.pkl ]; then
get_uniprot_sample \
--uniprot-fasta-path data/uniprot_sprot.fasta \
--output-path "data/sampled_id_2_seq.pkl" \
--sample-size 10000 > outputs/logs/swissprot_sampling.log 2>&1
else
echo "data/sampled_id_2_seq.pkl exists already. You might want to stash it before re-writing the file by the sampling script."
fiAlso, for experimental (wet-lab) validation, we sample Swiss-Prot for negative examples with the same script, while ensuring that the sampled sequences are not present in the training set.
cd TerpeneMiner
conda activate terpene_miner
if [ ! -f data/sampled_id_2_seq_experimental.pkl ]; then
get_uniprot_sample \
--uniprot-fasta-path data/uniprot_sprot.fasta \
--output-path "data/sampled_id_2_seq_experimental.pkl" \
--blacklist-path "data/sampled_id_2_seq.pkl" \
--sample-size 1000 > outputs/logs/swissprot_sampling_experimental.log 2>&1
else
echo "data/sampled_id_2_seq_experimental.pkl exists already. You might want to stash it before re-writing the file by the sampling script."
ficd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.data_preparation.cleaning_data_from_raw_tps_tableThis data preprocessing script is application-specific. It would require a separate implementation for other enzyme families. For that reason, the script is not configurable via command line arguments.
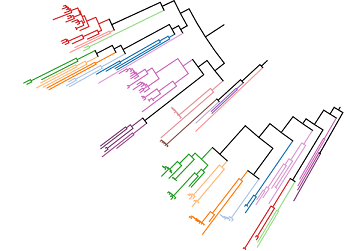
To check the generalization of our models to novel TPS sequences, we need to ensure that groups of similar sequences always stay either in train or in test fold. We construct a phylogenetic tree of our cleaned TPS dataset to compute groups of similar sequences. Clades of the tree define the groups of similar sequences. E.g., in the following visualization of our TPS phylogenetic subtree, the clade-based groups have the same color:
We share the computed phylogenetic groups in data/phylogenetic_clusters.pkl for reproducibility.
To compute a clade-based sequence group on your own, run
cd TerpeneMiner
conda activate terpene_miner
if [ ! -f data/phylogenetic_clusters.pkl ]; then
get_phylogeny_based_clusters \
--tps-cleaned-csv-path data/TPS-Nov19_2023_verified_all_reactions.csv \
--n-workers 64 > outputs/logs/phylogenetic_clusters.log 2>&1
else
echo "data/phylogenetic_clusters.pkl exists already. You might want to stash it before re-writing the file using the script for phylogenetic-tree-based sequence clustering."
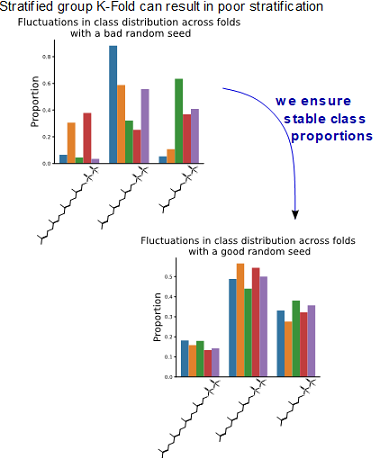
fiWe use 5-fold cross-validation (CV) for performance assessment. As described above, we ensure that similar sequences end
up
the same fold. Technically, we validate via group 5-fold CV. To ensure stable validation scores across folds,
we stratify based on the TPS substrate. As default StratifiedGroupKFold implementation from sklearn.model_selection
can result in class imbalance, we implement an iterative splitting procedure by varying random seeds and selecting the
one with the best correspondence of class proportions between folds (the proportion correspondence is compared using
Jensen–Shannon divergence).
We share the computed folds in data/tps_folds_nov2023.h5 for reproducibility.
To compute the folds on your own, run
cd TerpeneMiner
conda activate terpene_miner
if [ ! -f data/tps_folds_nov2023.h5 ]; then
python -m terpeneminer.src.data_preparation.get_balanced_stratified_group_kfolds \
--negative-samples-path data/sampled_id_2_seq.pkl \
--tps-cleaned-csv-path data/TPS-Nov19_2023_verified_all_reactions.csv \
--n-folds 5 \
--split-description stratified_phylogeny_based_split_with_minor_products \
> outputs/logs/kfold_with_minors.log 2>&1
else
echo "data/tps_folds_nov2023.h5 exists already. You might want to stash it before re-writing the file using the script for stratified group k-fold computation."
fi
Then, to store the folds in corresponding CSVs, run
cd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.data_preparation.store_folds_into_csv \
--negative-samples-path data/sampled_id_2_seq.pkl \
--tps-cleaned-csv-path data/TPS-Nov19_2023_verified_all_reactions.csv \
--kfolds-path data/tps_folds_nov2023.h5 \
--split-description stratified_phylogeny_based_split_with_minor_products \
> outputs/logs/kfold_with_minors_to_csv.log 2>&1For the majority of proteins, AlphaFold2(AF2)-predicted structures can be downloaded using the following script from my ProFun library. If you ran the installation steps, then the ProFun library is already installed and you can use the following command:
cd TerpeneMiner
conda activate terpene_miner
awk -F, '$1 != "" && $1 != "\"" && $1 != "Uniprot ID" {print $1}' "data/TPS-Nov19_2023_verified_all_reactions_with_neg_with_folds.csv" | sort | uniq > tps_ids.txt
alphafold_struct_downloader \
--path-to-file-with-ids tps_ids.txt \
--structures-output-path "data/alphafold_structs" \
--n-jobs 64
rm tps_ids.txtThe downloaded structures will be stored in the data/alphafold_structs folder. For the remaining few without precomputed AF2 prediction,
one of the easiest ways to run AF2 is by
using ColabFold [5] by
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S and Steinegger M.).
For reproducibility, we share the AF2 predictions for the sequences without DeepMind-precomputed AF2 predictions on zenodo as alphafold_additional.zip. You can simply add its contents to the data/alphafold_structs
folder and run the consequent evaluation steps.
Also, for illustration purposes, we store AF2 predictions for the archaeal TPSs we discovered in the
folder data/alphafold_structs on GitHub. There, we also put there a randomly selected TPS with UniProt accession B9GSM9, and PDBe structures we used for domain
standards.
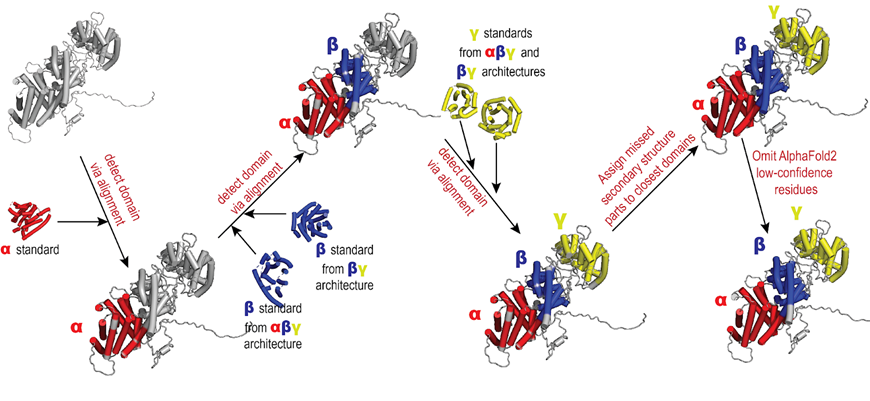
A high-level overview of our pipeline for TPS structure segmentation into domains is depicted in the following figure:
To use the algorithms for segmenting AF2 structures into TPS-specific domains, run
cd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.structure_processing.domain_detections \
--needed-proteins-csv-path "data/TPS-Nov19_2023_verified_all_reactions_with_neg_with_folds.csv" \
--input-directory-with-structures "data/alphafold_structs/" \
--n-jobs 16 --detections-output-path "data/filename_2_detected_domains_completed_confident.pkl" \
--store-domains --domains-output-path "data/detected domains" > outputs/logs/tps_structures_segmentation.log 2>&1To perform pairwise comparison of the detected domains with the use our alignment-based algorithms, run
cd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.structure_processing.compute_pairwise_similarities_of_domains \
--name all \
--n-jobs 64 \
--precomputed-scores-path "data/precomputed_tmscores.pkl" > outputs/logs/pairwise_comparisons.log 2>&1Note the --precomputed-scores-path argument. It is used to store the previously computed TM-scores.
For the efficiency of any future extensions of the project, we share the precomputed TM-scores in data/precomputed_tmscores.pkl on GitHub.
Also note, that if you have access to more servers, you might want to load-balance the pairwise comparison computation across your machines as shown below:
# Number of machines to split the workload across
n_machines = 15
# Total number of regions, i.e. the detected structural domains, to process
regions_total = len(regions_completed_confident_all)
# Calculate the delta value, which determines how many pairs each machine will process
# Overall, we need to fill an upper-triangular distance matrix of size regions_total x regions_total
delta = regions_total**2 / 2 // n_machines + 1
# Initialize counters
start_i = 0 # Keeps track of the current index in the pairwise comparison
start_prev = 0 # Keeps track of the previous start index for comparison
split_indices = [0] # List to hold the indices where splits across machines will occur
# Loop over each region to calculate the split indices
for i in range(regions_total):
for j in range(i + 1, regions_total):
start_i += 1 # Increment start_i for each pair (i, j)
# If the difference between the current and previous start index exceeds delta, record a split
if start_i - start_prev >= delta:
split_indices.append(i)
start_prev = start_i
# Append the total number of regions to the split indices to cover the last segment
split_indices.append(regions_total)
def print_script(i: int, split_indices: list[int]=split_indices):
"""
Print the command to process a segment of the regions.
Parameters:
- i (int): The index in split_indices that determines the start and end of the segment.
- split_indices (list of int): The list of indices where the workload is split.
"""
print(
f"""python -m terpeneminer.src.structure_processing.compute_pairwise_similarities_of_domains --start-i {split_indices[i]} --end-i {split_indices[i + 1]} --n-jobs 64 --name all
"""
)
# Loop over each segment and print the corresponding script command
for i in range(len(split_indices) - 1):
print_script(i)For convenience, we share all the raw pairwise comparison results in data/tps_domains_and_comparisons.zip, which are
subsequently used for domain clustering.
For clustering, run
cd TerpeneMiner
jupyter notebookThen, execute the notebook notebooks/notebook_3_clustering_domains.ipynb.
First, we extract protein-language-model's (PLM's) embeddings.
cd TerpeneMiner
conda activate terpene_miner
. scripts/extract_all_embeddings.sh > outputs/logs/embeddings_extraction.log 2>&1Parameters of the models and/or hyperparameter search can be modified in configs.
cd TerpeneMiner
conda activate terpene_miner
terpene_miner_main run > outputs/logs/models_training.log 2>&1This command will automatically retrieve all models specified in the configs folder.
If you want to exclude some model, put .ignore suffix to the corresponding folder in configs.
If you want to run a single model, run
cd TerpeneMiner
conda activate terpene_miner
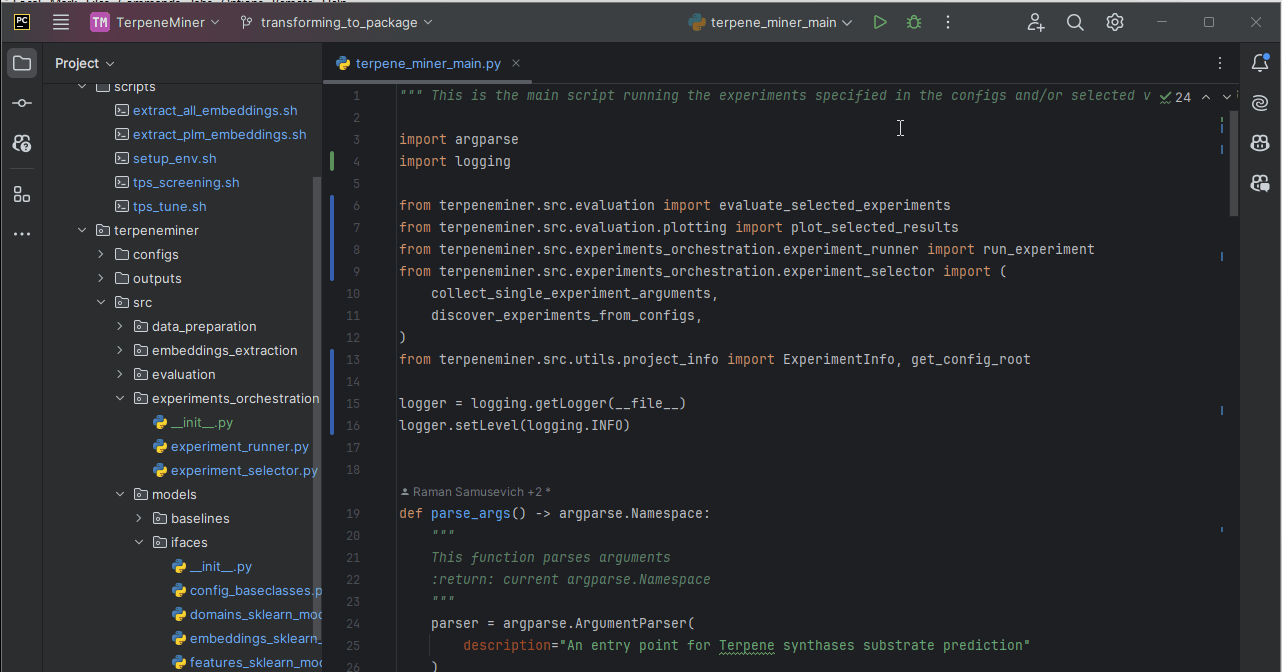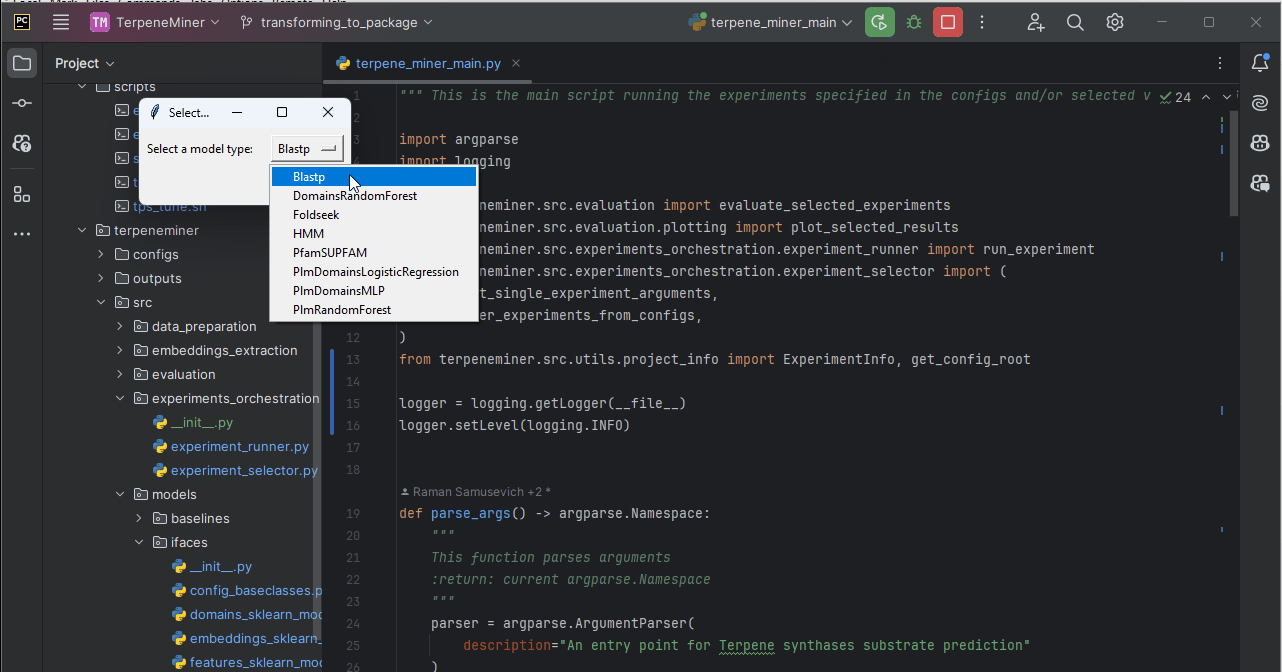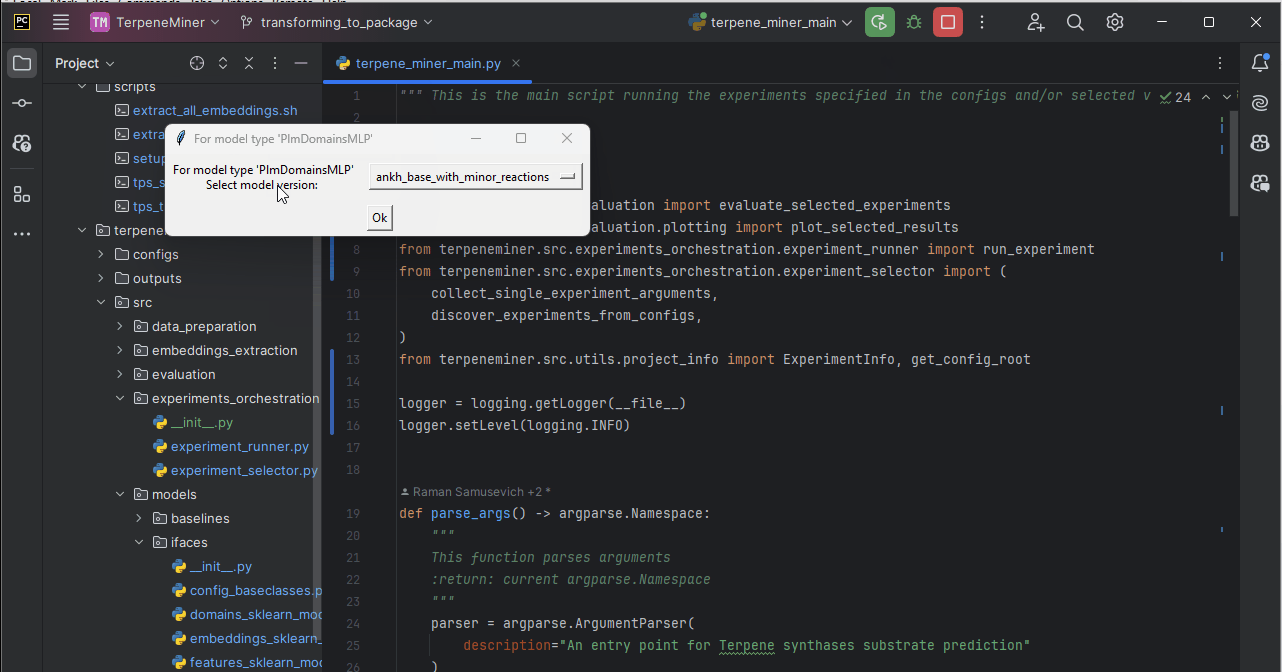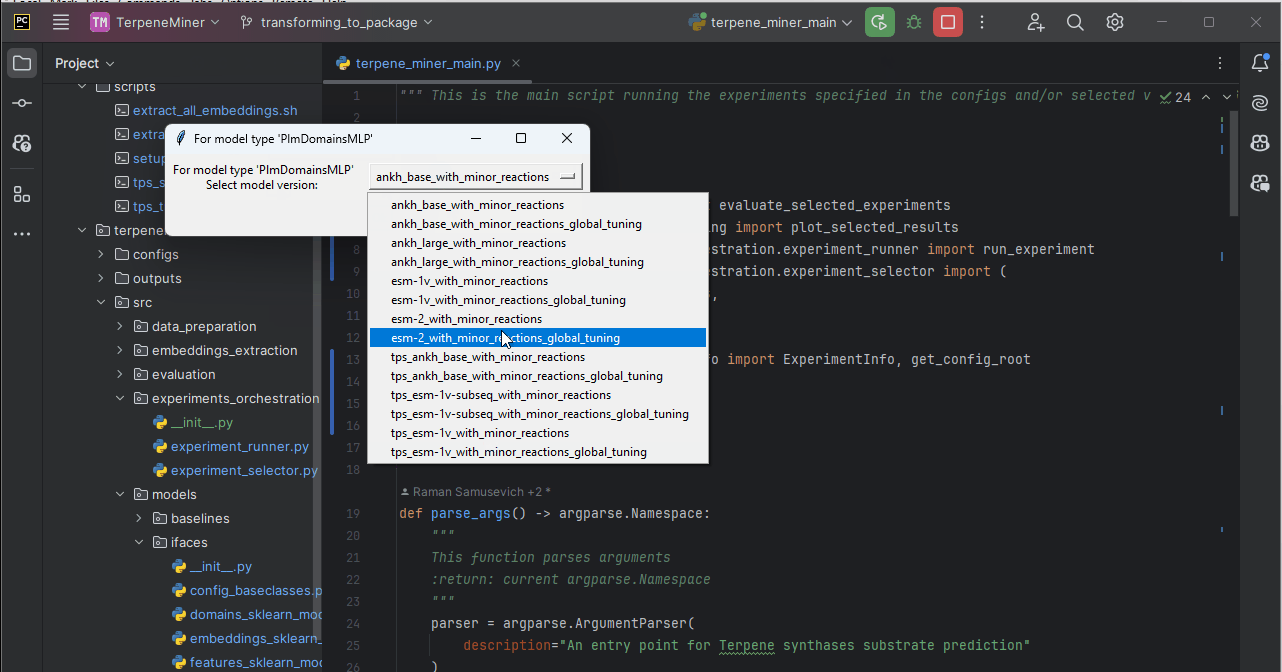
terpene_miner_main --select-single-experiment runOn headless servers, you would be prompted to select one of the available configs via the command line:

Otherwise, you can select a model via a simple GUI.
After training a PlmDomainsRandomForest, to select the most important domains for the best-performing model, run
cd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.models.plm_domain_faster.get_domains_feature_importances \
--top-most-important-domain-features-per-model 200 --output-path "data/domains_subset.pkl" > outputs/logs/domains_subset.log 2>&1-
Please note, that if you run into error
FileNotFoundError: [Errno 2] No such file or directory: '<path>/model_fold_0.pkl', you might need to re-run the training of the model while specifying thesave_trained_model: truein the config. -
In case of troubles, download outputs of the hyperparameter optimization from zenodo as
outputs.zipand unzip its contents to theoutputsfolder. Then the end-to-end derivation of the most important domains can be achieved with the following commands:
cd TerpeneMiner
conda activate terpene_miner
# training the model using pre-computed hyperparameters: select PlmDomainsRandomForest
terpene_miner_main --select-single-experiment run --load-hyperparameters
# gather the most important domains
python -m terpeneminer.src.models.plm_domain_faster.get_domains_feature_importances \
--top-most-important-domain-features-per-model 200 --output-path "data/domains_subset.pkl" --use-all-foldsAlso, for the sake of reproducibility, we share the selected domains in data/domains_subset.pkl on GitHub.
#### 5 - Parallelized hyperparameter optimization
If you want to run hyperparameter optimization in parallel, you can use the following:
```bash
cd TerpeneMiner
conda activate terpene_miner
bash scripts/tps_tune.sh # see the script for more details and accommodate to your use caseFor reproducability, we share outputs of the hyperparameter optimization
on zenodo as outputs.zip. You can simply unzip its contents to the outputs
folder and run the consequent evaluation steps.
If you want to train a single model using the best hyperparameters found during the previously run optimization, then set optimize_hyperparams: false in the config and run
cd TerpeneMiner
conda activate terpene_miner
terpene_miner_main --select-single-experiment run --load-hyperparametersIf you then want to gather the corresponding checkpoints into an easy-to-use pickle file, run
python -m terpeneminer.src.screening.gather_classifier_checkpoints --output-path data/classifier_domain_and_plm_checkpoints.pkl --use-all-foldsTo evaluate all configured models, run
cd TerpeneMiner
conda activate terpene_miner
terpene_miner_main evaluateAgain, if you want to evaluate a single model, run
cd TerpeneMiner
conda activate terpene_miner
terpene_miner_main --select-single-experiment evaluate --output-filename single_model_specific_nameand select the experiment you are interested in.
To evaluate detection of the TPSs, run
terpene_miner_main evaluate --classes "isTPS" --output-filename tps_detection
terpene_miner_main evaluate --classes "isTPS" --id-2-category-path data/id_2_kingdom_dataset.pkl --output-filename tps_detection_per_kingdom
To evaluate separately for individual kingdoms, run
terpene_miner_main evaluate --id-2-category-path data/id_2_kingdom_dataset.pkl --output-filename per_kingdomFinally, to evaluate results separately per entries with and without Pfam/SUPFAM/InterPro protein signatures, run
terpene_miner_main evaluate --id-2-category-path data/id_2_domains_presence.pkl --output-filename per_interpro_signaturesOnce the performance evaluation is done, you can visualize the results.
- To visualize main evaluation results, run
terpene_miner_main visualizeIt will generate the following set of plots in the outputs/evaluation_results
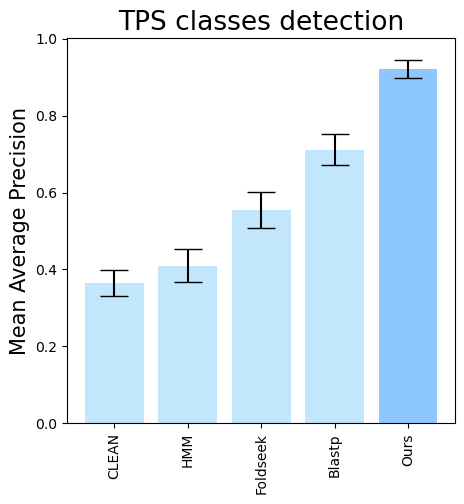
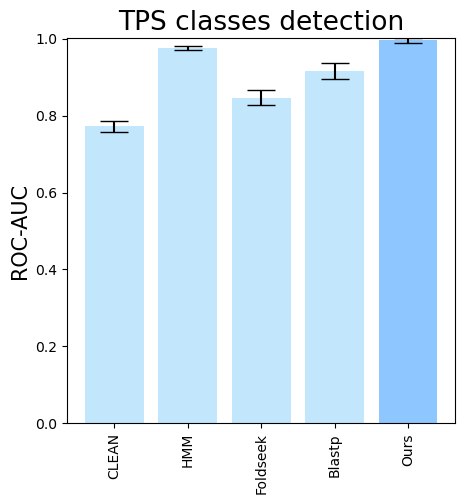
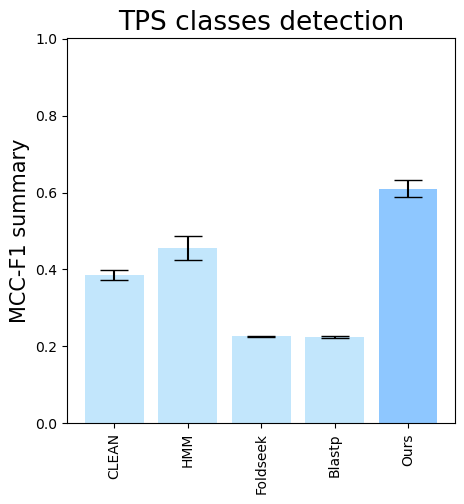
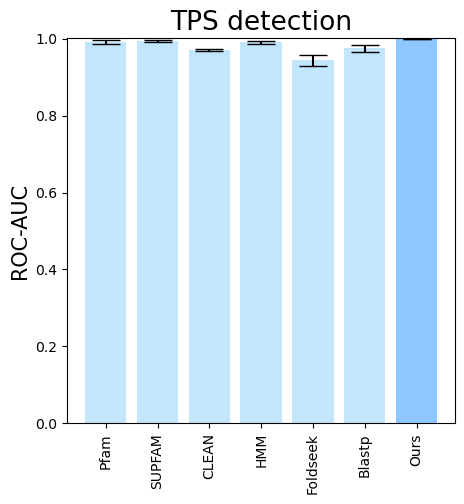
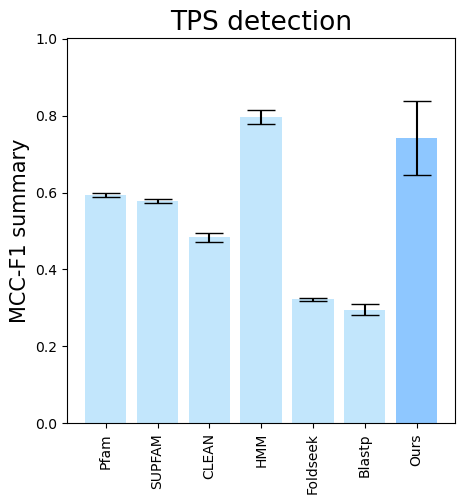
folder (all_results_Mean Average Precision.png, all_results_ROC-AUC.png, all_results_MCC-F1 summary.png):
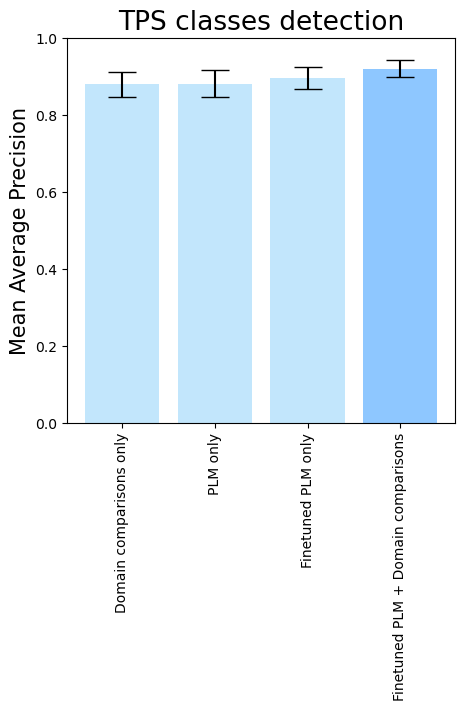
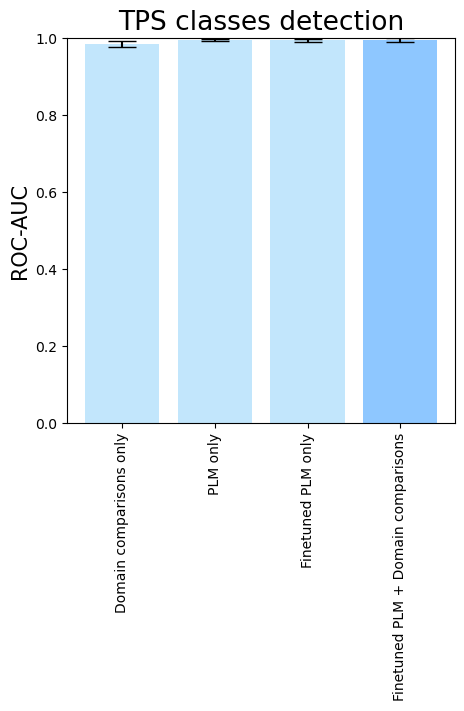
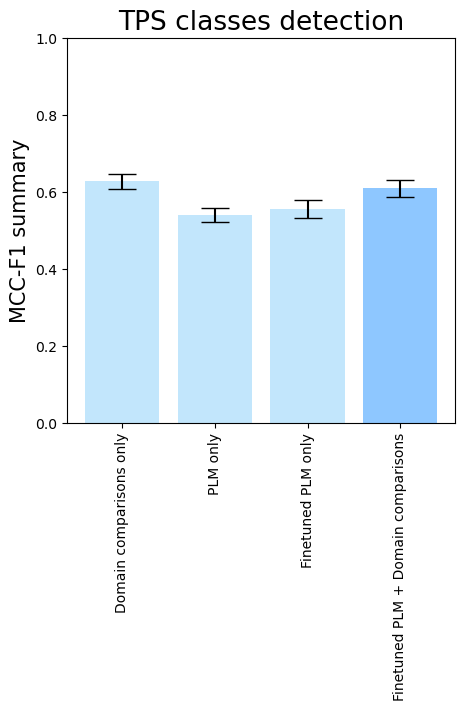
- To see the isolated importance of domain-comparison features, of PLM embeddings, and PLM fine-tuning, run
terpene_miner_main visualize --models \
DomainsRandomForest__with_minor_reactions_global_tuning PlmRandomForest__esm-1v_with_minor_reactions_global_tuning PlmRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset \
--model-names "Domain comparisons only" "PLM only" "Finetuned PLM only" "Finetuned PLM + Domain comparisons"\
--subset-name "ablation_study"Then, the following plots will be generated in the outputs/evaluation_results
folder (ablation_study_Mean Average Precision.png, ablation_study_ROC-AUC.png, ablation_study_MCC-F1 summary.png):
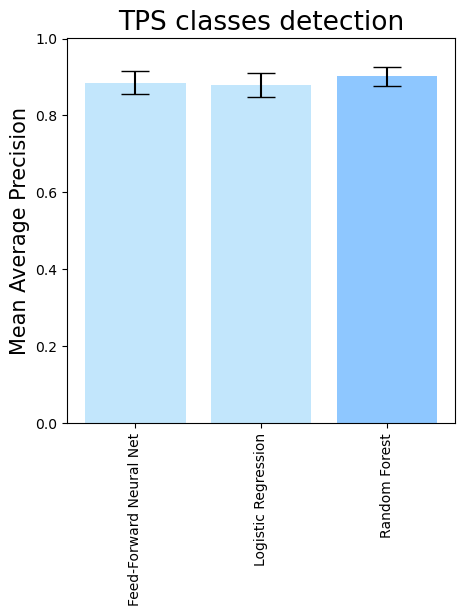
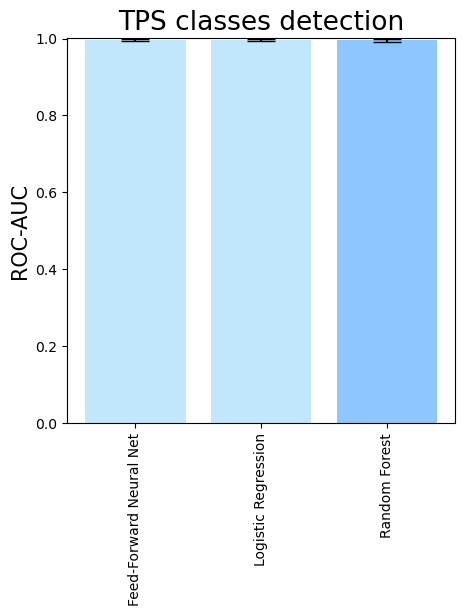
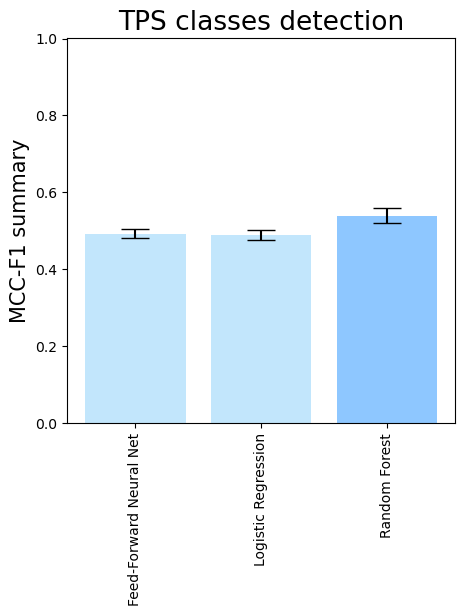
- To compare different downstream classifiers on top of the same features (PLM embeddings + domain comparisons), run
terpene_miner_main visualize --models \
PlmDomainsMLP__tps_esm-1v-subseq_with_minor_reactions_global_tuning PlmDomainsLogisticRegression__tps_esm-1v_with_minor_reactions_global_tuning PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning\
--model-names "Feed-Forward Neural Net" "Logistic Regression" "Random Forest"\
--subset-name "different_models_best_feats"Then, the following plots will be generated in the outputs/evaluation_results
folder (different_models_best_feats_Mean Average Precision.png, different_models_best_feats_ROC-AUC.png, different_models_best_feats_MCC-F1 summary.png):
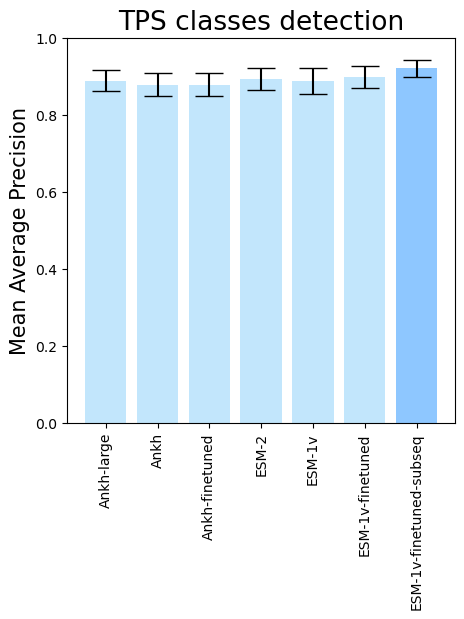
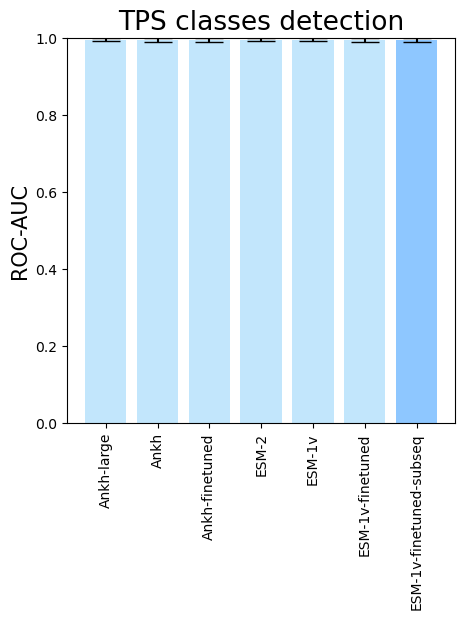
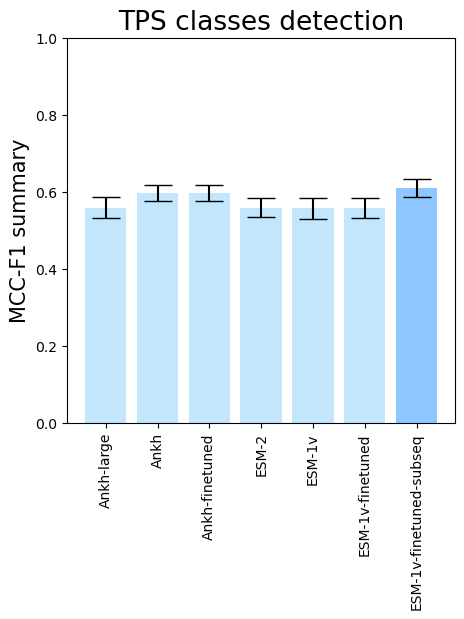
- To see the performance for different PLMs, run
terpene_miner_main visualize --models \
PlmDomainsRandomForest__ankh_large_with_minor_reactions_global_tuning PlmDomainsRandomForest__ankh_base_with_minor_reactions PlmDomainsRandomForest__tps_ankh_base_with_minor_reactions PlmDomainsRandomForest__esm-2_with_minor_reactions_global_tuning PlmDomainsRandomForest__esm-1v_with_minor_reactions_global_tuning PlmDomainsRandomForest__tps_esm-1v_with_minor_reactions_global_tuning PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names Ankh-large Ankh Ankh-finetuned ESM-2 ESM-1v ESM-1v-finetuned ESM-1v-finetuned-subseq \
--subset-name "random_forest_different_plm"Then, the following plots will be generated in the outputs/evaluation_results
folder (random_forest_different_plm_Mean Average Precision.png, random_forest_different_plm_ROC-AUC.png, random_forest_different_plm_MCC-F1 summary.png):
The following plots will be generated in the outputs/evaluation_results
folder (all_results_Average Precision_per_type.png, all_results_ROC AUC_per_type.png, all_results_MCC-F1 summary_per_type.png):
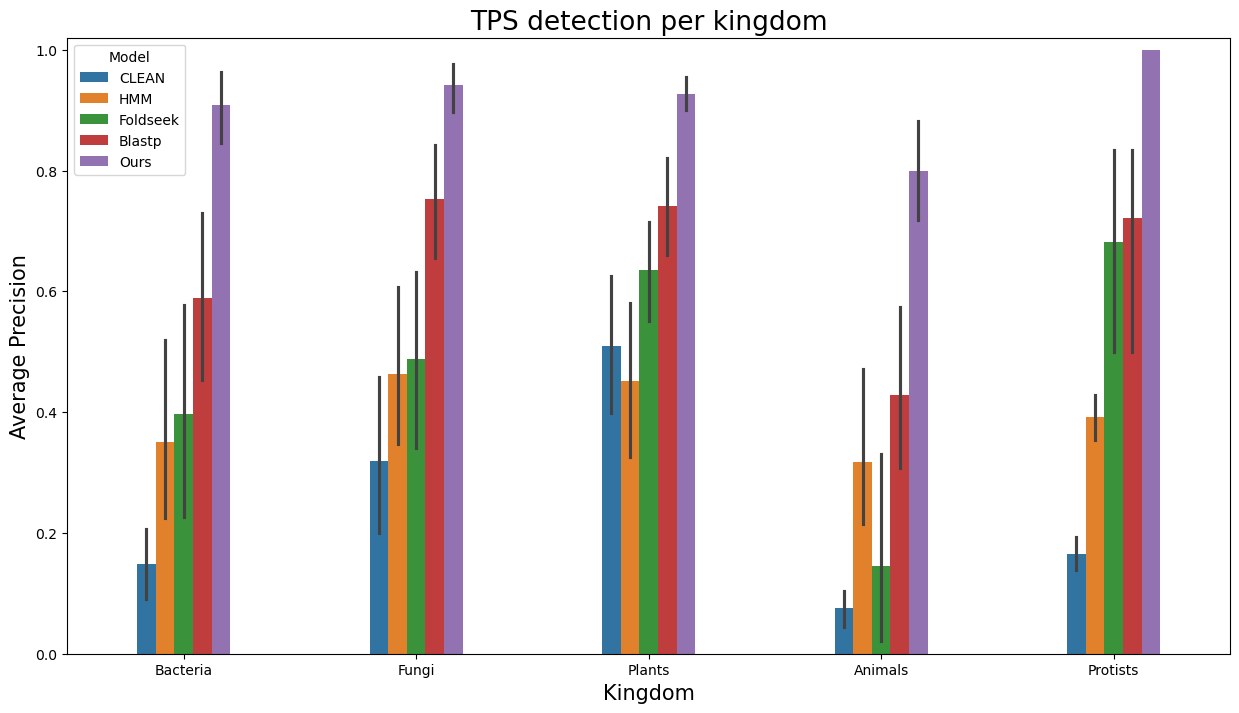
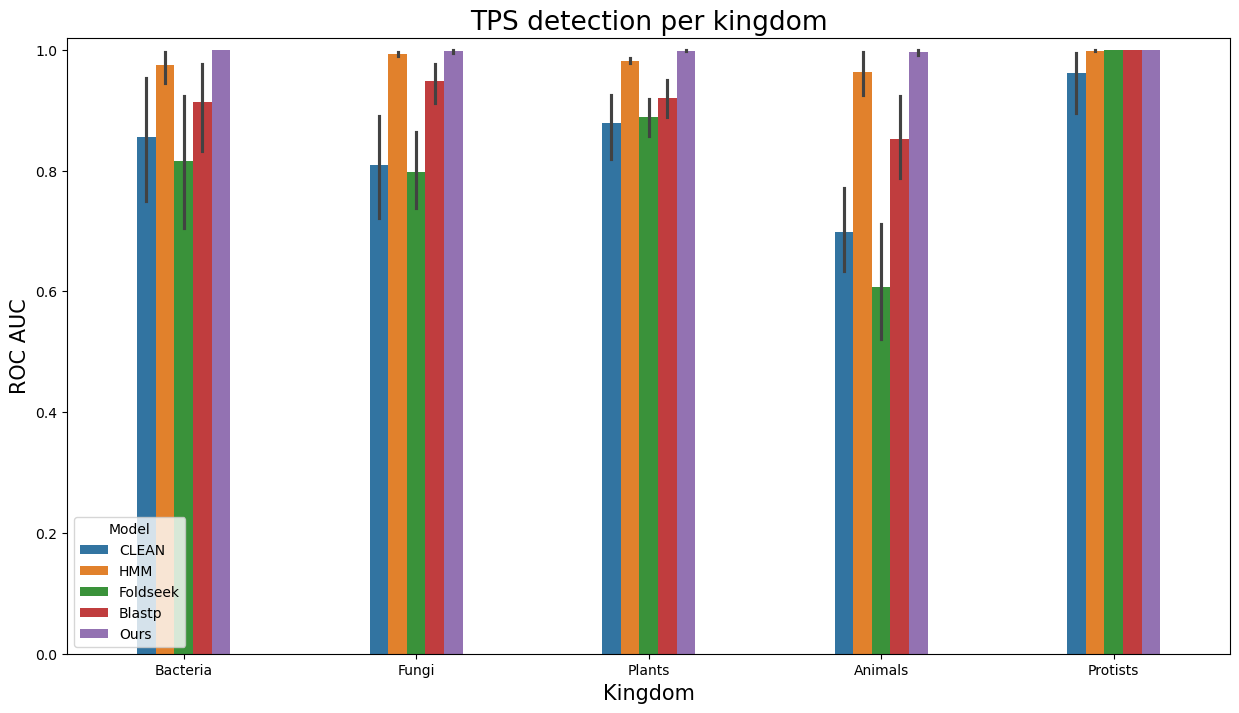
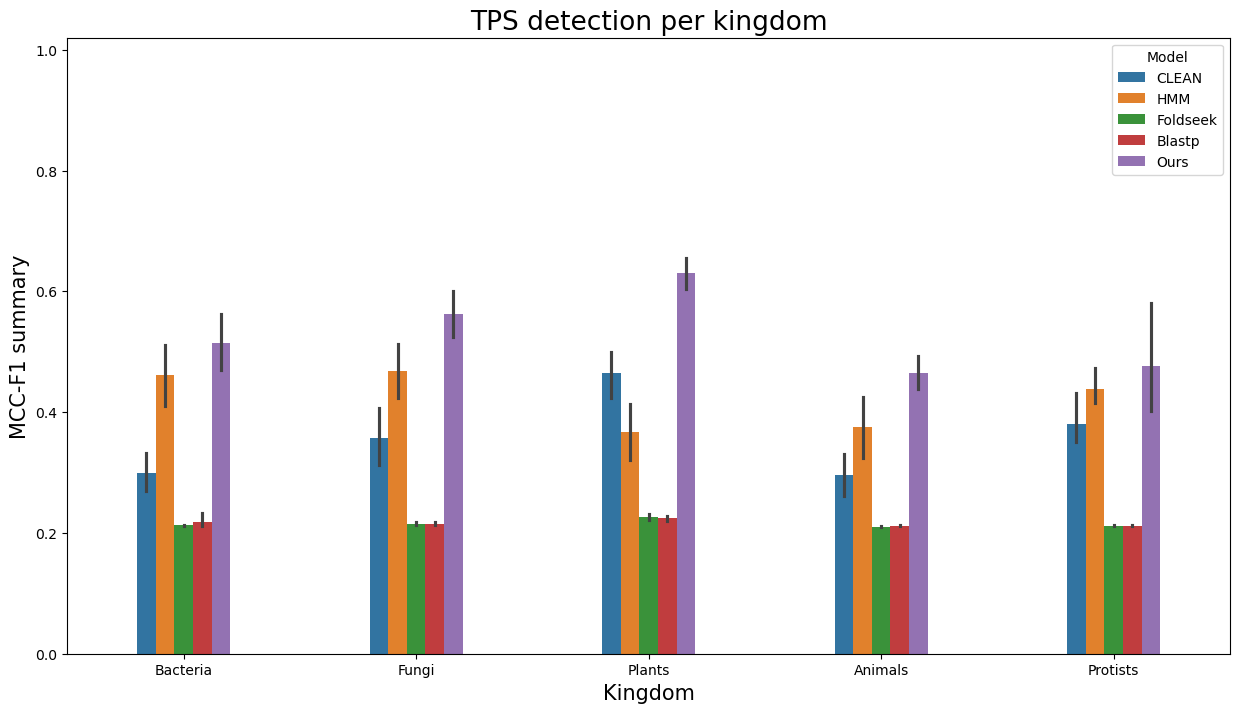
- Similarly, to visualize performance separately per each kingdom, run
terpene_miner_main visualize --plot-barplots-per-category --models \
CLEAN__with_minor_reactions HMM__with_minor_reactions Foldseek__with_minor_reactions Blastp__with_minor_reactions PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names "CLEAN*" HMM Foldseek Blastp Ours \
--category-name Taxon --id-2-category-path data/id_2_kingdom_dataset.pkl --eval-output-filename per_kingdom \
--categories-order Bacteria Fungi Plants Animals Protists VirusesThe following plots will be generated in the outputs/evaluation_results
folder (per_kingdom_Average Precision_per_kingdom.png, per_kingdom_ROC AUC_per_kingdom.png, per_kingdom_MCC-F1 summary_per_kingdom.png):
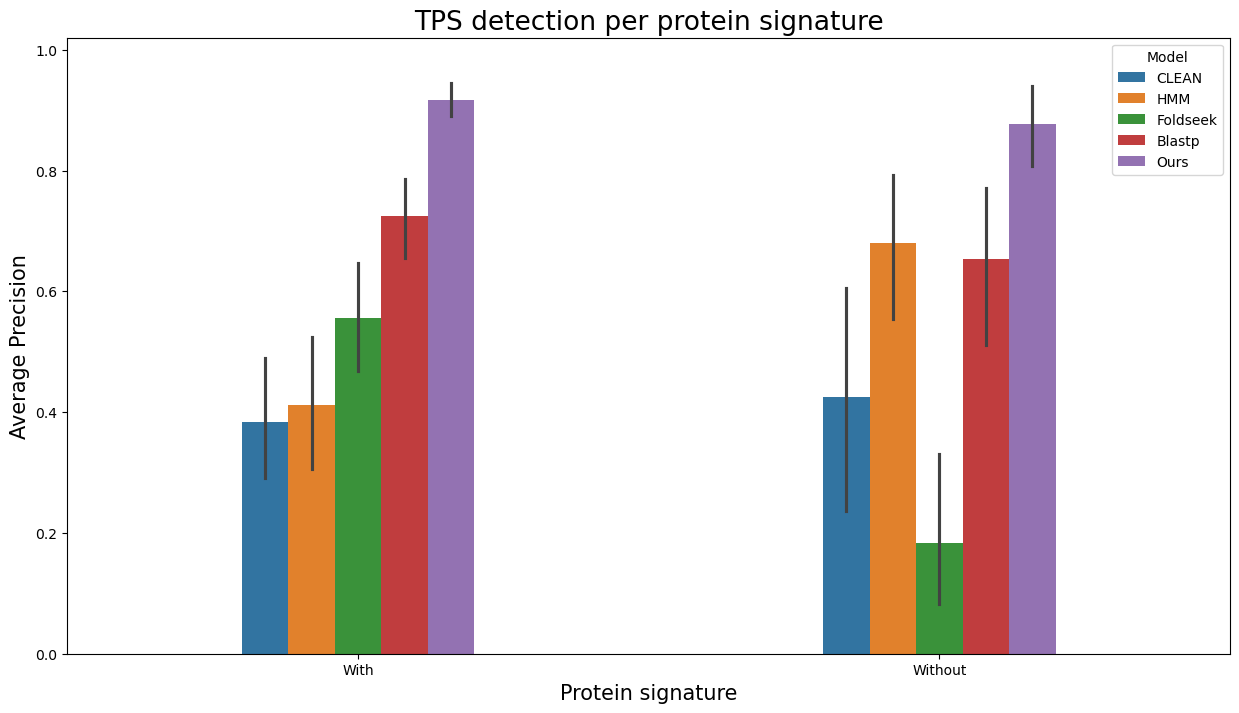
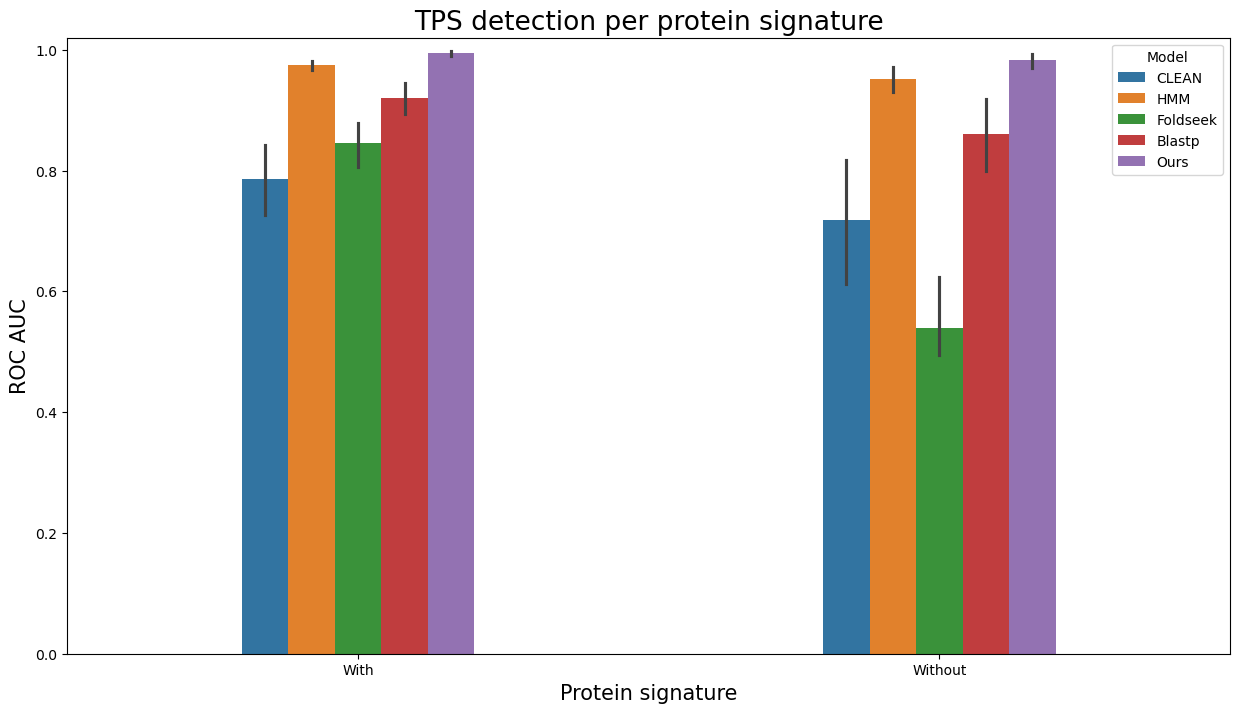
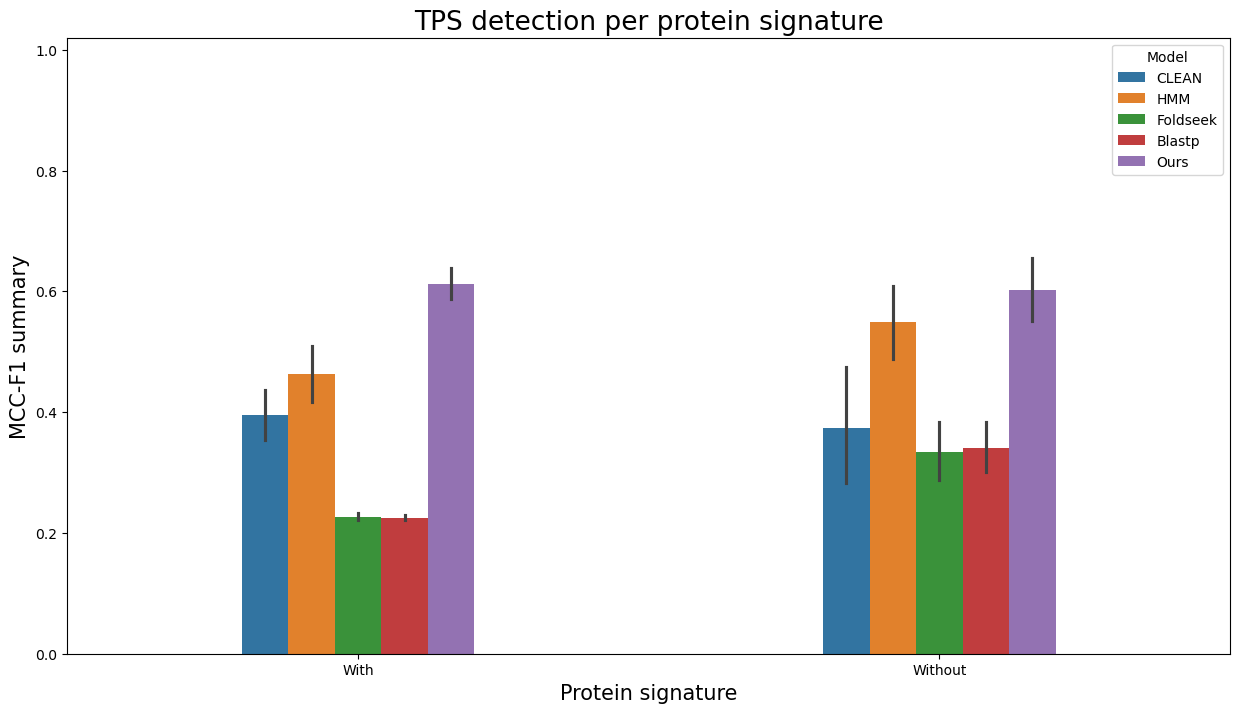
Analogously, to visualize evaluation results as barplots separately per entries with and without Pfam/SUPFAM/InterPro protein signatures, run
terpene_miner_main visualize --eval-output-filename all_results --plot-barplots-per-category --models \
CLEAN__with_minor_reactions HMM__with_minor_reactions Foldseek__with_minor_reactions Blastp__with_minor_reactions PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names "CLEAN*" HMM Foldseek Blastp Ours \
--category-name "Protein signature" --id-2-category-path data/id_2_domains_presence.pkl --eval-output-filename per_interpro_signatures \
--categories-order With Without
The following plots will be generated in the outputs/evaluation_results
folder (
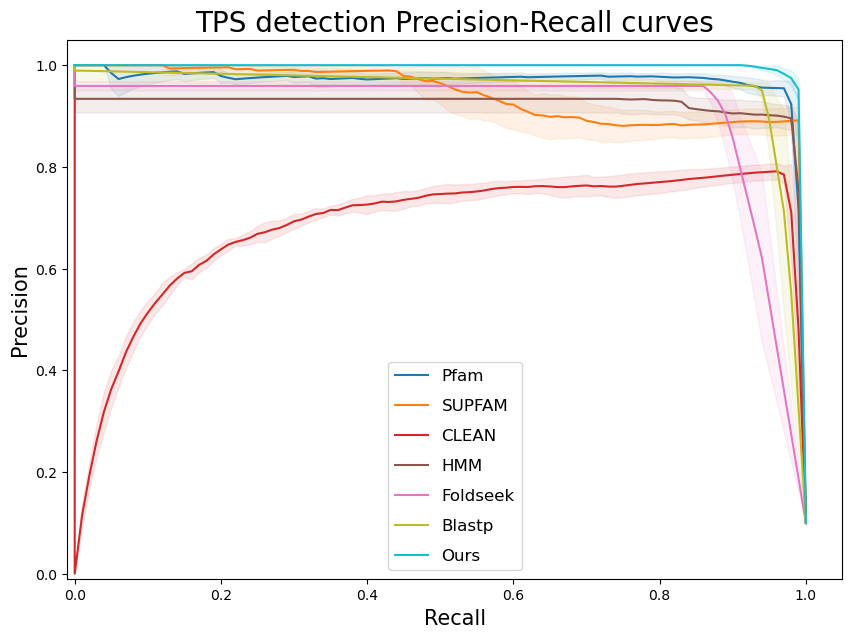
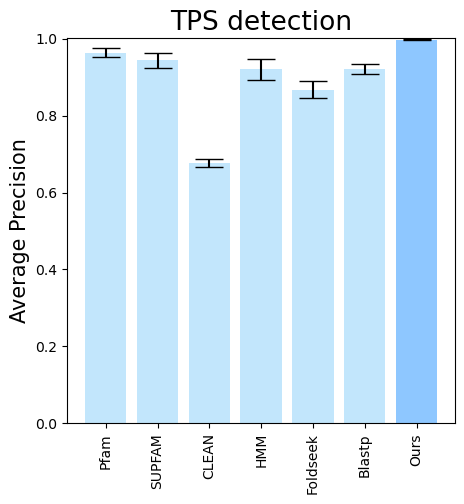
terpene_miner_main visualize --eval-output-filename tps_detection --plot-tps-detection --models \
CLEAN__with_minor_reactions Foldseek__with_minor_reactions Blastp__with_minor_reactions HMM__with_minor_reactions PfamSUPFAM__supfam PfamSUPFAM__pfam PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names "CLEAN*" Foldseek Blastp HMM SUPFAM Pfam Oursterpene_miner_main visualize --plot-barplots-per-category --models \
CLEAN__with_minor_reactions Foldseek__with_minor_reactions Blastp__with_minor_reactions HMM__with_minor_reactions PfamSUPFAM__supfam PfamSUPFAM__pfam PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names "CLEAN*" Foldseek Blastp HMM SUPFAM Pfam Ours --category-name Taxon --id-2-category-path data/id_2_kingdom_dataset.pkl --eval-output-filename tps_detection_per_kingdom \
--categories-order Bacteria Fungi Plants Animals Protists Viruses- To visualize performance per different TPS types, run
terpene_miner_main visualize --eval-output-filename tps_detection --plot-boxplots-per-type --models \
CLEAN__with_minor_reactions Foldseek__with_minor_reactions HMM__with_minor_reactions Blastp__with_minor_reactions PfamSUPFAM__supfam PfamSUPFAM__pfam PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names "CLEAN*" Foldseek Blastp HMM SUPFAM Pfam Ours This will generate the following plots outputs/evaluation_results/tps_detection_*:
This is a global mean across all TPSs. So basically, it is mainly the performance on major classes. To see the performance for different TPS types, run commands like the following:
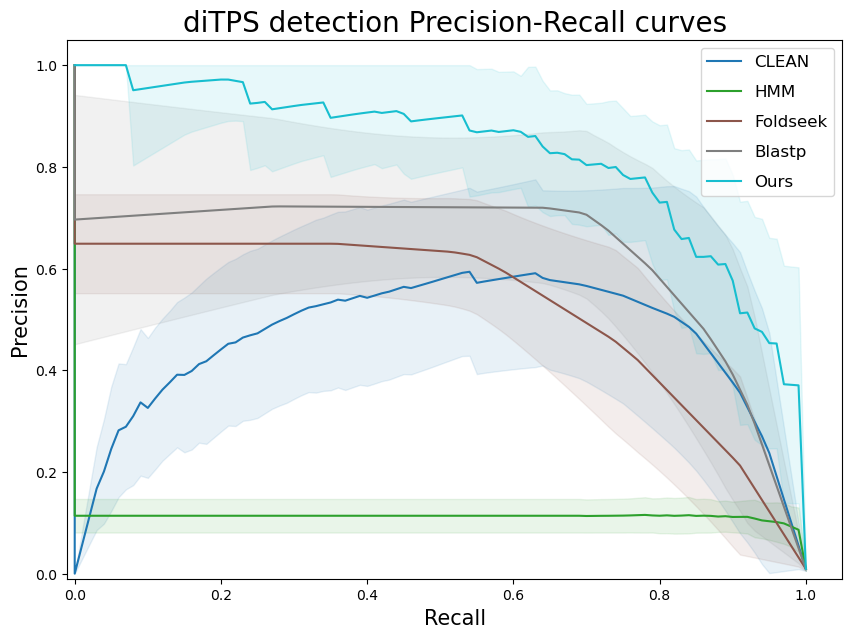
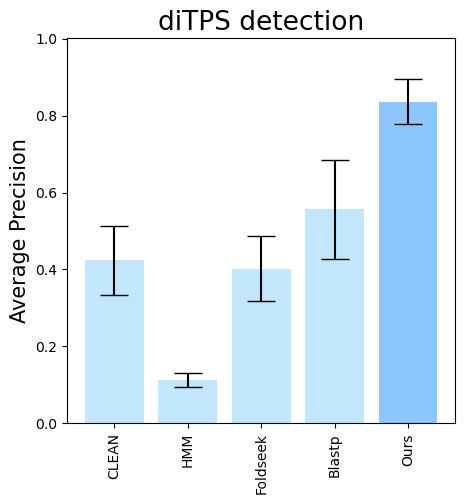
terpene_miner_main visualize --eval-output-filename all_results --plot-tps-detection --models \
CLEAN__with_minor_reactions HMM__with_minor_reactions Foldseek__with_minor_reactions Blastp__with_minor_reactions PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names "CLEAN*" HMM Foldseek Blastp Ours \
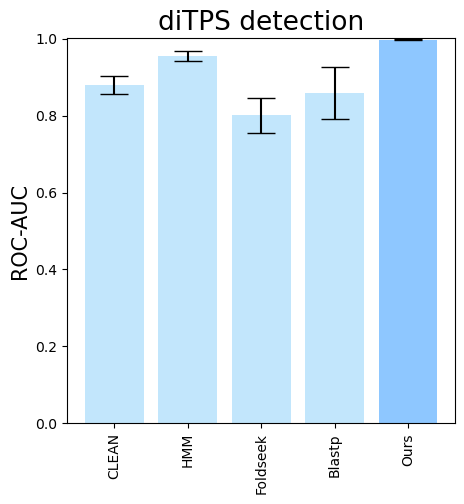
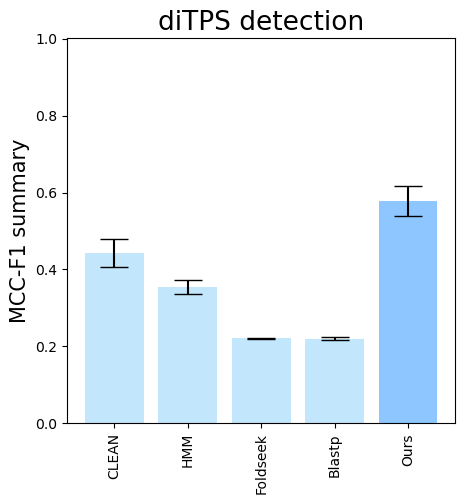
--subset-name "di_detection" --type-detected di
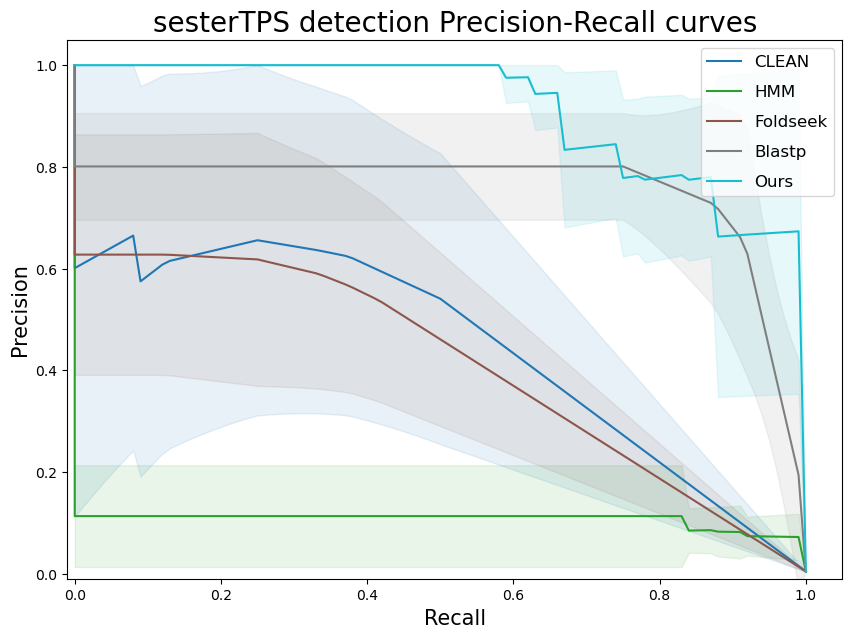
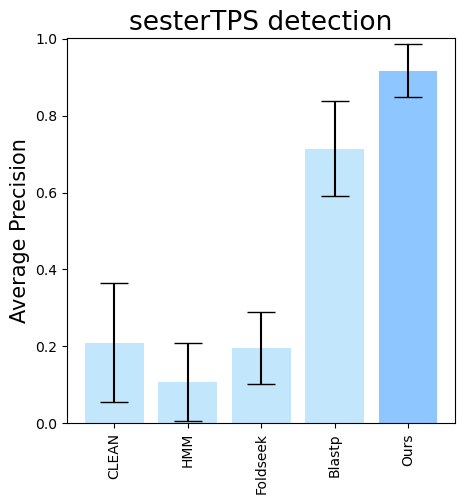
terpene_miner_main visualize --eval-output-filename all_results --plot-tps-detection --models \
CLEAN__with_minor_reactions HMM__with_minor_reactions Foldseek__with_minor_reactions Blastp__with_minor_reactions PlmDomainsRandomForest__tps_esm-1v-subseq_with_minor_reactions_global_tuning_domains_subset\
--model-names "CLEAN*" HMM Foldseek Blastp Ours \
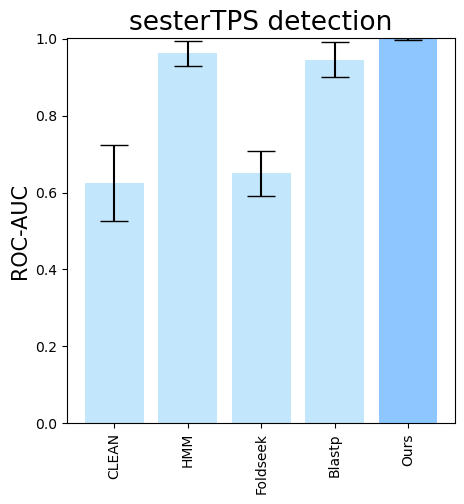
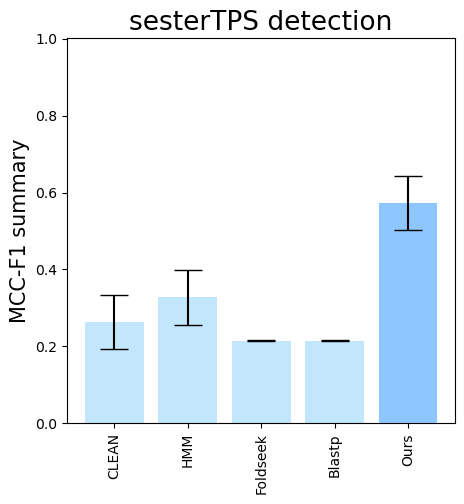
--subset-name "sester_detection" --type-detected sester
Before screening large databases, you need to gather the trained models. To do so, run
cd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.screening.gather_classifier_checkpoints --output-path data/classifier_checkpoints.pklNext, to estimate the required number of workers for the screening, run
cd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.screening.estimate_number_of_workers --fasta-path data/uniprot_trembl.fasta --delta 40000 --n-gpus 8Note that delta stands for the number of sequences to be processed by a single GPU on a worker.
To screen large databases, then run
sbatch --array=0-<number_of_workers> scripts/tps_screening.sh "data/uniprot_trembl.fasta" "trembl_screening_output" 0.4where <number_of_workers> is the number of workers estimated in the previous step. Please note, that you might have no
slurm on your cluster,
and you would need to set up the cluster environment yourself.
This will store individual hits as separate files. To merge them into a single CSV file, run
cd TerpeneMiner
conda activate terpene_miner
python -m terpeneminer.src.screening.gather_detections_to_csv --screening-results-root "trembl_screening/detections_plm" --output-path "trembl_screening/detections_plm/detections_first_batch.csv" --delete-individual-filesSamusevich, R., Hebra, T. et al. Highly accurate discovery of terpene synthases powered by machine learning reveals functional terpene cyclization in Archaea. bioRxiv ( 2024). https://doi.org/10.1101/2024.01.29.577750
@article{samusevich2024tps,
title={Highly accurate discovery of terpene synthases powered by machine learning reveals functional terpene cyclization in Archaea},
author={Samusevich, Raman and Hebra, Teo and Bushuiev, Roman and Bushuiev, Anton and {\v{C}}alounov{\'a}, Tereza and Smr{\v{c}}kov{\'a}, Helena and Chatpatanasiri, Ratthachat and Kulh{\'a}nek, Jon{\'a}{\v{s}} and Perkovi{\'c}, Milana and Engst, Martin and Tajovsk{\'a}, Ad{\'e}la and others},
journal={bioRxiv},
pages={2024--01},
year={2024},
publisher={Cold Spring Harbor Laboratory}