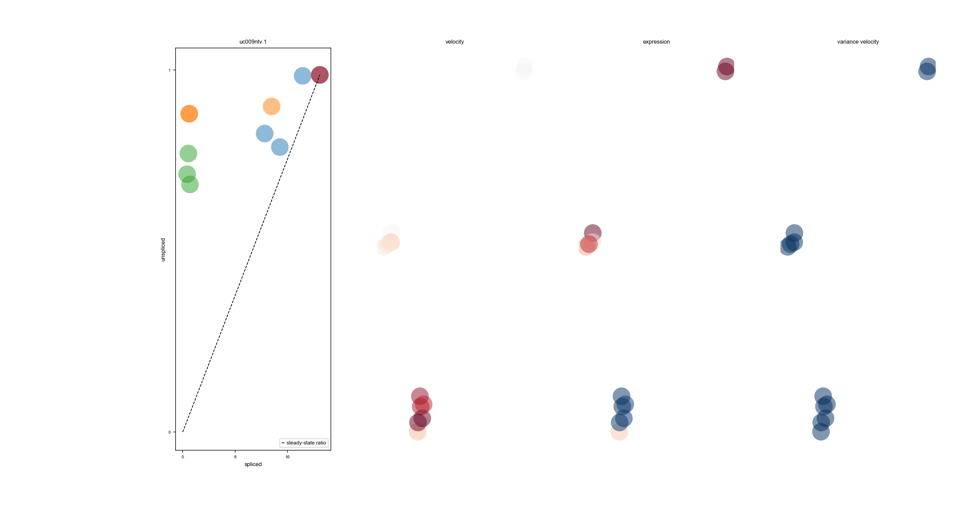
RNA-Velocity in simple words the ratio of spliced vs unspliced transcripts. mRNA splicing kinetics: Transcriptional induction for a particular gene results in an increase of (newly transcribed) precursor unspliced mRNAs while, conversely, repression or absence of transcription results in a decrease of unspliced mRNAs. Please see: https://www.nature.com/articles/s41586-018-0414-6)
Source: https://scvelo.readthedocs.io/
Please suggest improvements by forking a branch and report bugs on Issues section.
STAR [version 2.6.1 =>] [https://github.com/alexdobin/STAR]
python3 [version 3.7.1] [https://www.python.org/downloads/]
R [version 3.5.0 =>] [https://www.r-project.org/]
simplesam - a python module [https://simplesam.readthedocs.io/en/latest/#]
samtools [version 1.8.0 =>] [http://www.htslib.org/]
velocyto [http://velocyto.org/velocyto.py/install/index.html]
scVelo [https://scvelo.readthedocs.io/]
loompy - a python module [http://linnarssonlab.org/loompy/installation/index.html]
scanpy [https://scanpy.readthedocs.io/en/stable/installation.html]
pandas - a python module [https://pandas.pydata.org/]
numpy - a python module [https://numpy.org/]
matplotlib - a python module [https://matplotlib.org/]
- Map the raw RNA-Seq reads to genome of interest using STAR
- Sort and Index the Bam file resulting from Step1 using samtools
- Run simplesam module in python to add UMI and Barcode tags
- Remove reads mapped to chr's (tagged as random) or short scaffolds using samtools
- Run Velocyto on the bam resulting from Step4 with -U option
- Run scVelo on the loom file resulting from Step5
STAR --genomeDir STAR-Genome-Index --readFilesIn FASTQ-FILES --readFilesCommand gunzip -c
--outFileNamePrefix OUTbam --runThreadN 4 --outSAMstrandField intronMotif --outSAMtype BAM Unsorted
--sjdbGTFfile Genome-GTF
samtools sort OUTbamAligned.out.bam -o OUTbamAligned.out.sorted.bam
samtools index OUTbamAligned.out.sorted.bam
Please see SimpleSam.py is available in the repository for download
python3 SimpleSam.py OUTbamAligned.out.sorted.bam OUTbamAligned.simplesam.sam
samtools view -b -o OUTbamAligned.simplesam.bam OUTbamAligned.simplesam.sam
samtools sort OUTbamAligned.simplesam.bam -o OUTbamAligned.simplesam.sorted.bam
samtools view -b OUTbamAligned.simplesam.sorted.bam chr1 chr10 chr11 chr12 chr13 chr14 chr15 chr16 chr17 chr18
chr19 chr20 chr21 chr22 chr2 chr3 chr4 chr5 chr6 chr7 chr8 chr9 chrM chrX chrY > OUTbamAligned.velocyto.temp.bam
samtools view -b -F 4 OUTbamAligned.velocyto.temp.bam > OUTbamAligned.velocyto.mapped.bam
samtools sort OUTbamAligned.velocyto.mapped.bam -o OUTbamAligned.velocyto.sorted.bam
samtools index OUTbamAligned.velocyto.sorted.bam
velocyto run -U -m GENOME-MASKED-GTF --outputfolder VELOCYTO_OUT OUTbamAligned.velocyto.sorted.bam Genome-GTF
If Multiple samples please combine .loom files using command -- python3 LoomCombine.py
Please see LoomCombine.py is available in the repository for download
python3 scVelo.py VELOCYTO_OUT.loom OUTFILE-PREFIX
scVelo.py has a requirement of Annotations.txt file to back-map sample information in case of multiple samples processed using combined loom file