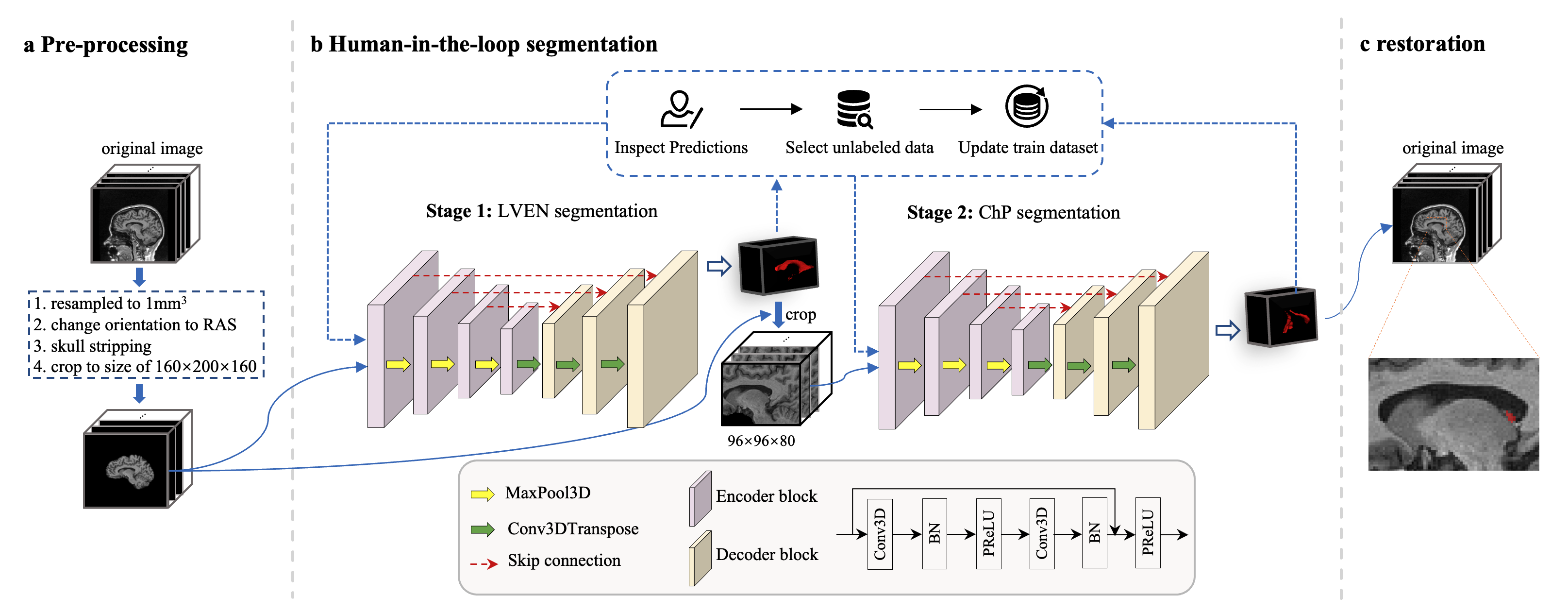
This is the pipeline which proposed in the paper published at Fluids and Barriers of the CNS. In this repository, we offer two methods to execute the ChP segmentation pipeline. One method involves directly executing the Python code, while the other method utilizes Docker.
- The following input forms are accepeted:
- Single NIfTI file (
.niior.nii.gz) - folder containing
.dcmseries - folder containing multiple NIfTI files
.txtfile where each row contains the NIfTI file path
- Output
Defualt:
resluts/, you can specify custom path.
Python (3.8.3). I believe Python 3.x should be fine, although I haven’t tested it on those versions.
git clone https://github.com/princeleeee/ChP-Seg.git
cd ChP-Seg
pip install -r requirements.txt
sed -i '1s/.*/import tensorflow.compat.v1 as tf/' /usr/local/lib/python3.8/site-packages/deepbrain/extractor.py # Necessary since Deepbrain is accomplished with Tensorlow 1.x
mkdir weights
# download deep learning models weights from https://drive.google.com/drive/folders/1M6fItRsPwV-hlww0YUdzabq9oz-RMNB0?usp=drive_link to weights folder.
python pipeline.py --input demo/I812923.nii.gz # This is a demo.Results/
├── file_collections.txt # all files input to the pipeline.
│
├── brain/ # save the results in the preprocessing and skull stripping stage.
│ │
│ ├── 0_resample/ # 1mm^3 RAS+ reorientation and resampled images save path.
│ │ ├── crop_range.txt
│ │ └── xxx.nii.gz
│ │
│ ├── 1_check/ # 3 plane .png check the crop option on 0_resample
│ ├── 1_err/ # 3 plane .png check the crop option on 0_resample
│ ├── 1_img/ # cropped skull stripped images from 0_resample, size: 160*200*160, crop range records is brain/0_resample/crop_range.txt
│ ├── 1_mask/ # brain mask on 1_img
│ │
│ └── 2_resample_inverse/ # restore images in 1_img to original image space.
│
├── ventricle/ # ventricle segmentation results folder
│ │
│ ├── 0_mask/ # segmentations resluts of lateral ventricles, size: 160*200*160
│ │
│ ├── 1_img_crop/ # crop brain/1_img to size 96*96*80
│ ├── 1_mask_crop/ # crop ventricle/0_mask to size 96*96*80
│ │
│ ├── 2_resampledT1_space/ # restore ventricle segmentation mask that matches brain/0_resample
│ │
│ ├── 3_orig_T1_space/ # restore ventricle segmentation mask that matches brain/2_resample_inverse
│ │
│ └── crop_range.txt # crop range records that generated ventricle/1_img_crop and ventricle/1_mask_crop
│
└── cp/ # choroid plexus segmentation resluts folder
│
├── 0_mask/ # segmentation results of choroid plexus, size 96*96*80
│
├── 1_mask_refine/ # refined segmentaiton results of cp/0_mask
│
├── 2_resampledT1_space/ # ChP segmentation match images in brain/0_resample
│
└── 3_orig_T1_space/ # ChP segmentation match images in brain/2_resample_inverse
# download deep learning models weights from https://drive.google.com/drive/folders/1M6fItRsPwV-hlww0YUdzabq9oz-RMNB0?usp=drive_link to a folder.
docker run -v $WEIGHTS_PATH_ON_HOST:/app/weights -v $OUTPUT_FOLDER_ON_HOST:/app/results -it --rm chp-seg bashIf you find our work helpful, please consider citing:
@article{li2024associations,
title={Associations between the choroid plexus and tau in Alzheimer’s disease using an active learning segmentation pipeline},
author={Li, Jiaxin and Hu, Yueqin and Xu, Yunzhi and Feng, Xue and Meyer, Craig H and Dai, Weiying and Zhao, Li and Alzheimer’s Disease Neuroimaging Initiative},
journal={Fluids and Barriers of the CNS},
volume={21},
number={1},
pages={56},
year={2024},
publisher={Springer}
}