Early accept at MICCAI 2022.
Adversarially Robust Prototypical Few-shot Segmentation with Neural-ODEs
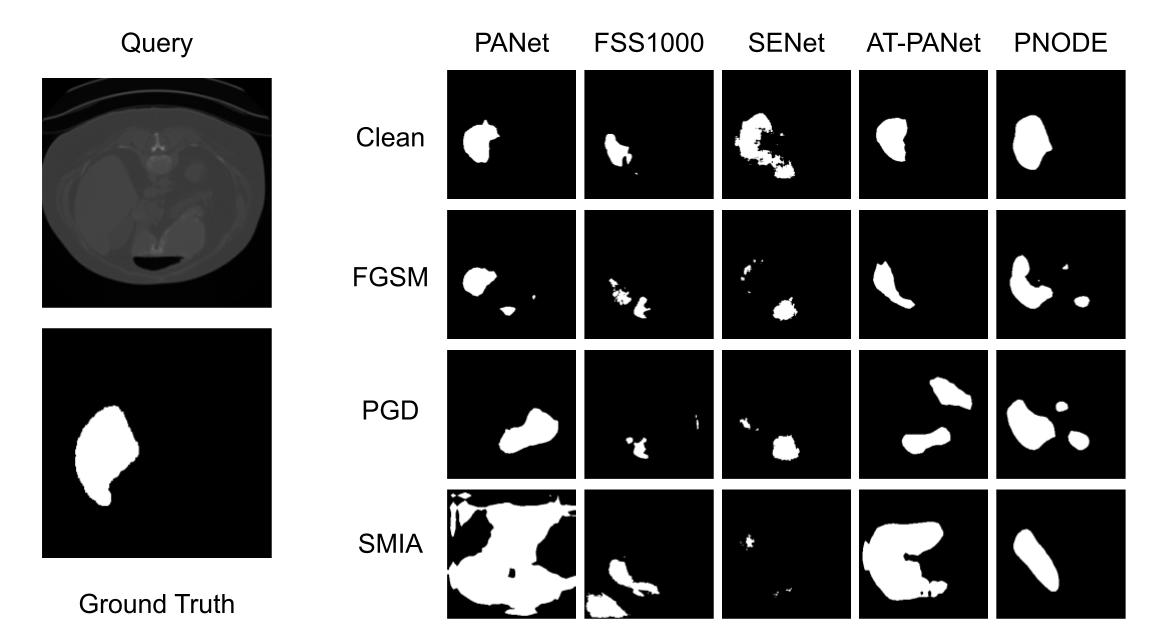
Predicted masks by different models for different attacks
To install this repository and its dependent packages, run the following.
git clone https://github.com/pnodemiccai/PNODE-FSS.git
cd PNODE-FSS
conda create --name PNODE-FSS # (optional, for making a conda environment)
pip install -r requirements.txt
Change the paths to BCV, CT-ORG and Decathlon datasets in config.py and test_config.py according to paths on your local. Also change the path to ImageNet pretrained VGG model weights in these files.
Trained model weights for PNODE, AT-PANet and all baselines on relevant settings can be found here.
Processed datasets of BCV, CT-ORG and Decathlon can be downloaded from here.
Implementations of baseline FSS methods used in this repository are adapted from here.
To train baseline methods, go to their respective folders and run
python3 train.py with model_name=<save-name> target=<test-target> n_shot=<shot>
For AT-PANet, run train_adversarial.py from PANet folder with the same arguments as before.
For PNODE, some extra arguments are needed. To train PNODE, run
python3 train.py with model_name=<save-name> target=<test-target> n_shot=<shot> ode_layers=3 ode_time=4
To train baseline methods, go to their respective folders and run
python3 test_attacked.py with snapshot=<weights-path> target=<test-target> dataset=<BCV/CTORG/Decathlon> attack=<Clean/FGSM/PGD/SMIA> attack_eps=<eps> to_attack=<q/s>
This command can be used for testing all models on all settings, namely 1-shot and 3-shot, liver and spleen and Clean, FGSM, PGD and SMIA with different epsilons.
BCV:
Liver: 6
Spleen: 1
CT-ORG:
Liver: 1
Decathlon:
Liver: 2
Spleen: 6