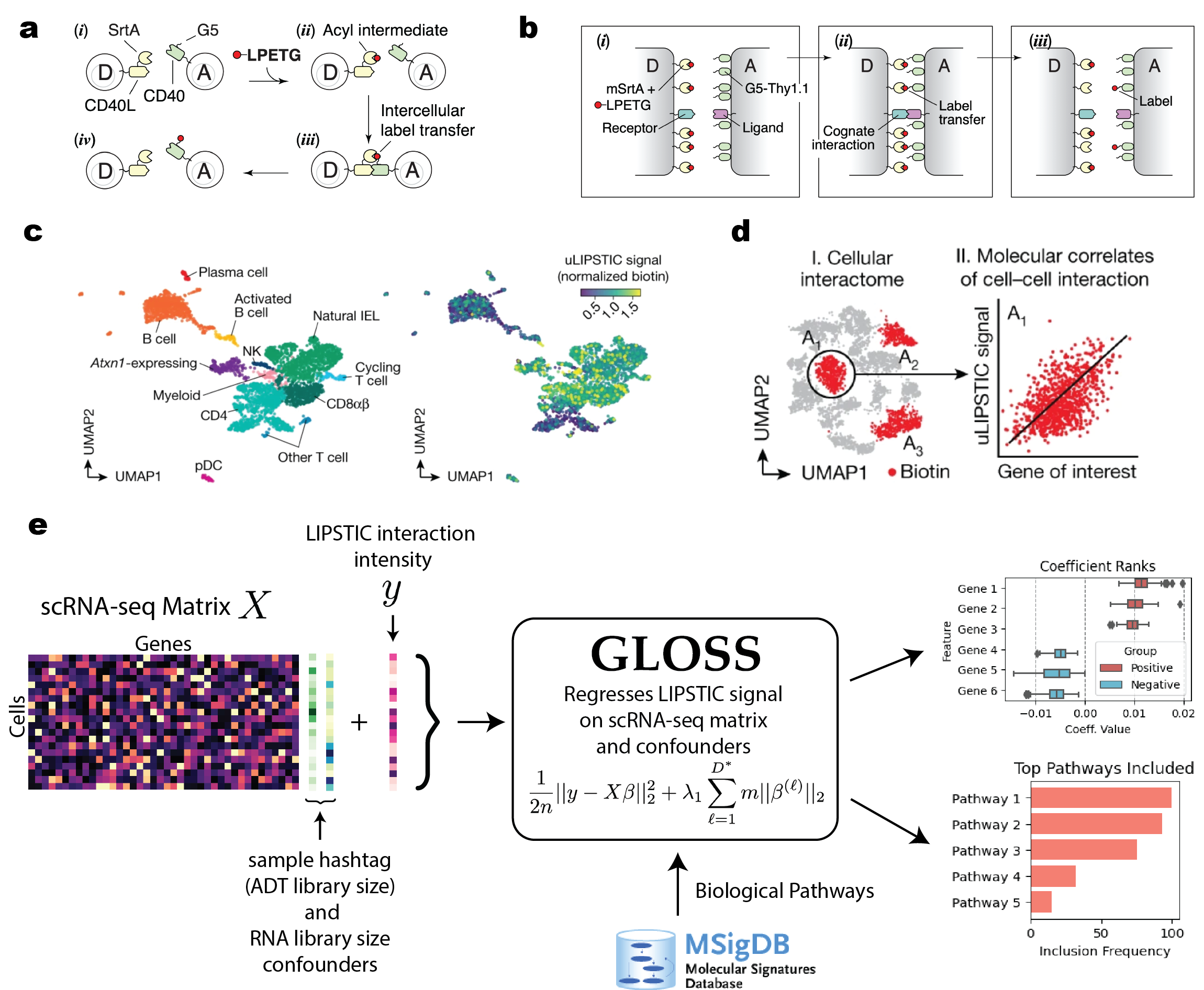
Gloss is a computational method that regresses LIPSTIC data on scRNA-seq features in paired LIPSTIC+scRNA-seq experiments. It accomplishes this by using an overlapping group lasso procedure using a set of pathways (which are preloaded into Gloss).
To install Gloss, simply run pip install . in this cloned repository in your environment.
Gloss assumes your data is annotated already, which you can specify under the resolutions field of the regression.
An example script of tuning Gloss using the Hallmark pathways on your data is shown below:
datapath = 'myscrna_anndata.h5ad' # with raw values in anndata.X
resolutions = {
'annotations' : ['Cell Type A', 'Cell Type B']
'annotations_fine' : ['Refined Cell Type A.1']
}
from Gloss.regresscv import RegressCV
regcv = RegressCV(datapath,
resolutions,
'hallmark',
'raw sample hashtag library size',
'RNA library size',
'uLIPSTIC raw signal')
Then, with the tuned parameters, you can run bootstrapped regression with Gloss to get robust estimates of the interaction-associated coefficients and pathways. To do this, here is an example script:
from Gloss.regressbootstrap import RegressBootstrap
regb = RegressBootstrap(datapath,
resolutions,
'hallmark',
myregcv.best_params,
100,
'raw sample hashtag library size',
'RNA library size',
'uLIPSTIC raw signal')
savepath = 'some bootstrap savepath'
import pickle
with open(savepath, 'wb') as handle:
pickle.dump(regb, handle, protocol=4)
Both of the above functions internally call the PrepData to preprocess the data and Regressor functions from Gloss to run the actual regression itself.
Gloss can also be used to separately normalize LIPSTIC signal, by regressing out the RNA library size and sample hashtag library size confounders at a dataset level. This is done by using Gloss to regress LIPSTIC signal only on the confounders, with no genes - this reduces to Lasso regression.
When PrepData is called, this is run by default, and the resulting normalized LIPSTIC expression is stored under the gloss_normalized_biotin observation field of the AnnData:
datapath = 'myscrna_anndata.h5ad' # with raw values in anndata.X
from Gloss.prepdata import PrepData
prepped_data = PrepData(datapath,
'hallmark',
'raw sample hashtag library size',
'RNA library size',
'uLIPSTIC raw signal')
prepped_data.adata.obs['gloss_normalized_biotin'] # normalized biotin stored here