With NSIDE = 32 the number of pixels is 12288. Using 3 Convnet layers with 32,16 and 8 filters respectively,I reduce the number of pixels to 48, then do a fully connected layer to 36. I decode it back the same way.
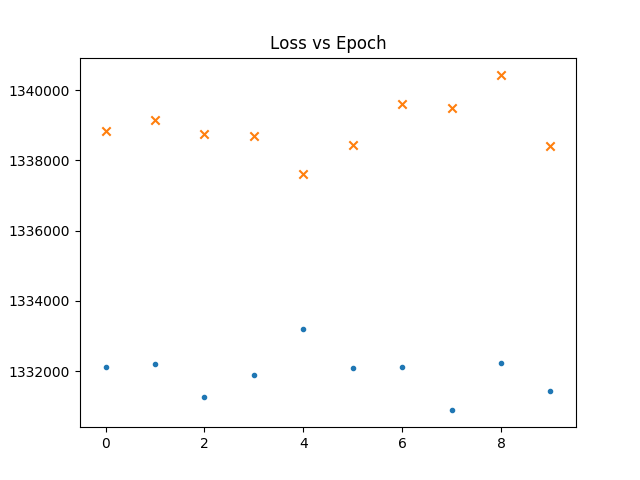
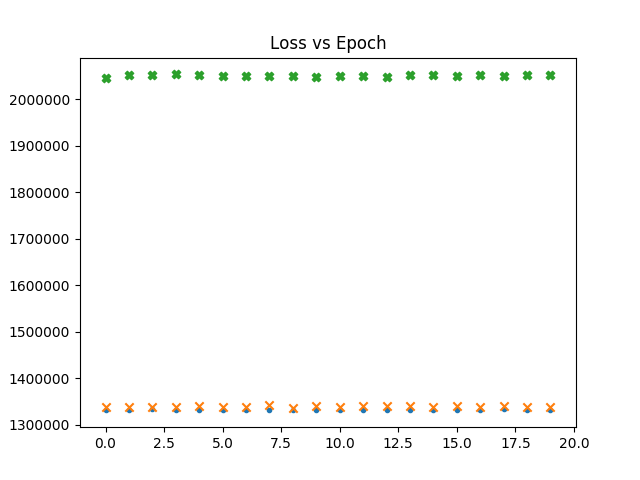
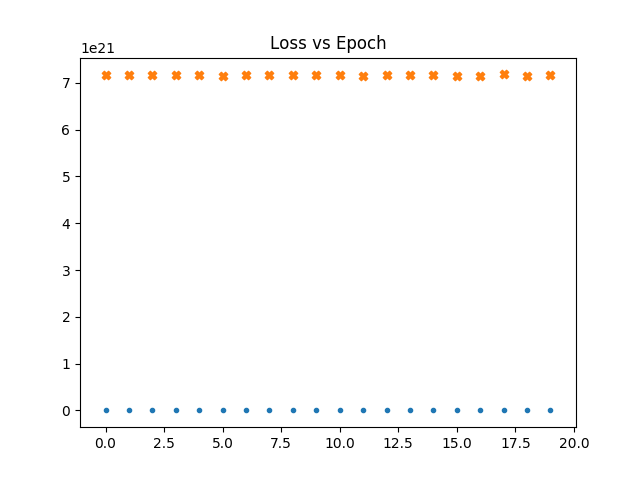
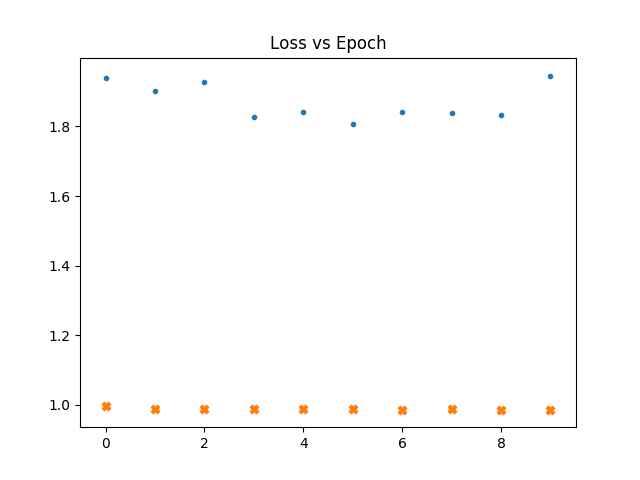
In the following graphs a dot represents the loss vs epochs for the gaussian maps. a cross for fnl = 1e-4. Thicker cross for fnl = 1e-3.
In the following plot the gaussian maps were generated after
calculating the power spectra of the non-gaussian maps, So that we
can make sure that the maps have the same power spectra
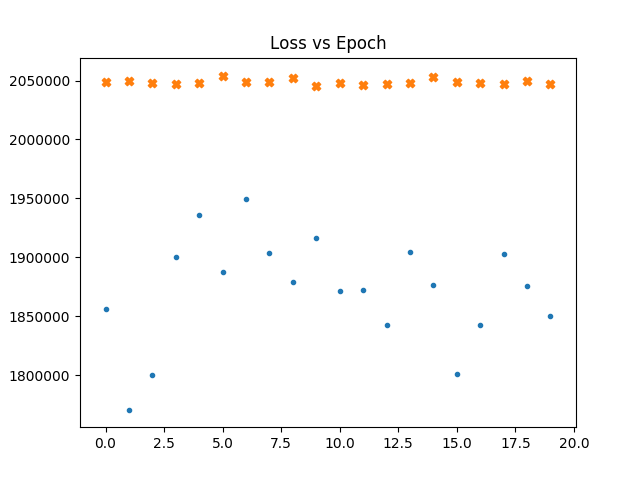
We see that even if we train with non gaussain maps. The loss doesn't go down.
 What is interesting is that though the network never seems to
learn the loss is already high for non gaussian maps.
What is interesting is that though the network never seems to
learn the loss is already high for non gaussian maps.
Maybe gaussian random initialization has something to do with it.
-
I need maps gaussian with different power spectra to test this incease in loss with non gaussianities further.
-
(More important) As of now, I am blindly reshaping the map array healpy gives me into a rectangle. I feel like spatial information is lost that way. I need a way to project it into a map and then get the two dimensional pixel data as an array.