Accompanies our paper:
Leonille Schweizer, Philipp Seegerer et al. "Analysing Cerebrospinal Fluid with Explainable Deep Learning: from Diagnostics to Insights", accepted for publication in Neuropathology and Applied Neurobiology.
The data are available at Zenodo.
The pipeline is built on Sacred, Snakemake and PyTorch.
First, create a local_config.py to set up Sacred:
log_dir_root = "/path/to/runs"
def setup_logger(ex):
# Configuration of Sacred experiment logger
def add_observers(ex):
# You need at least the FS observer because it creates the output dir where eg. model files and predictions are saved.
ex.observers.append(
sacred.observers.FileStorageObserver(log_dir_root)
)
# Add Mongo/Slack or other Sacred observers to the experiment
# E.g.
ex.observers.append(
sacred.observers.MongoObserver()
)
- Create an experiment configuration in YAML format, e.g.
yourconfig.yml( likedummy_experiment_config.yml), that contains the:experiment_name: name of the experiment in format<name>_v<version:02d>classes: list of classes that are included (long name, e.g. "aktivierter Lymphozyt")num_classes: number of class groups during training, i.e. output neuronscommon_config: dictionary of config updates common for all runs in this experiment.config_updates: dictionary of config updates for the individual runs, mapping"<run_id>" -> <config_update>.
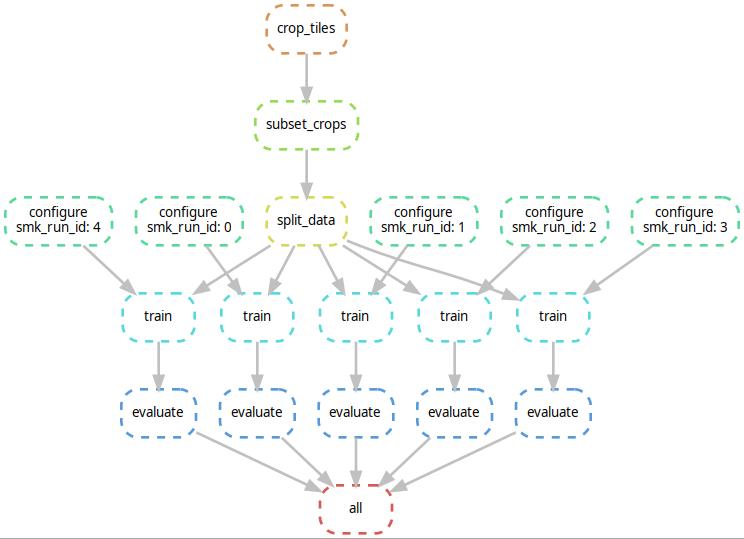
- Call the snakemake workflow
with
snakemake all --cores all --configfile yourconfig.yml. This will automatically detect which stages of the workflow have to be executed. Add your custom cluster configuration according to the snakemake docs. E.g., if--default-resources gpus=0 --cluster "qsub -l cuda={resources.gpus} -binding linear:4 -cwd -V -q all.q"is provided, distributes it to different qsub jobs (in parallel, if possible) . The stages are connected like (example for two experiment runs):
This will train the model and create a classification report
in /path/to/runs/<sacred_run_id>/