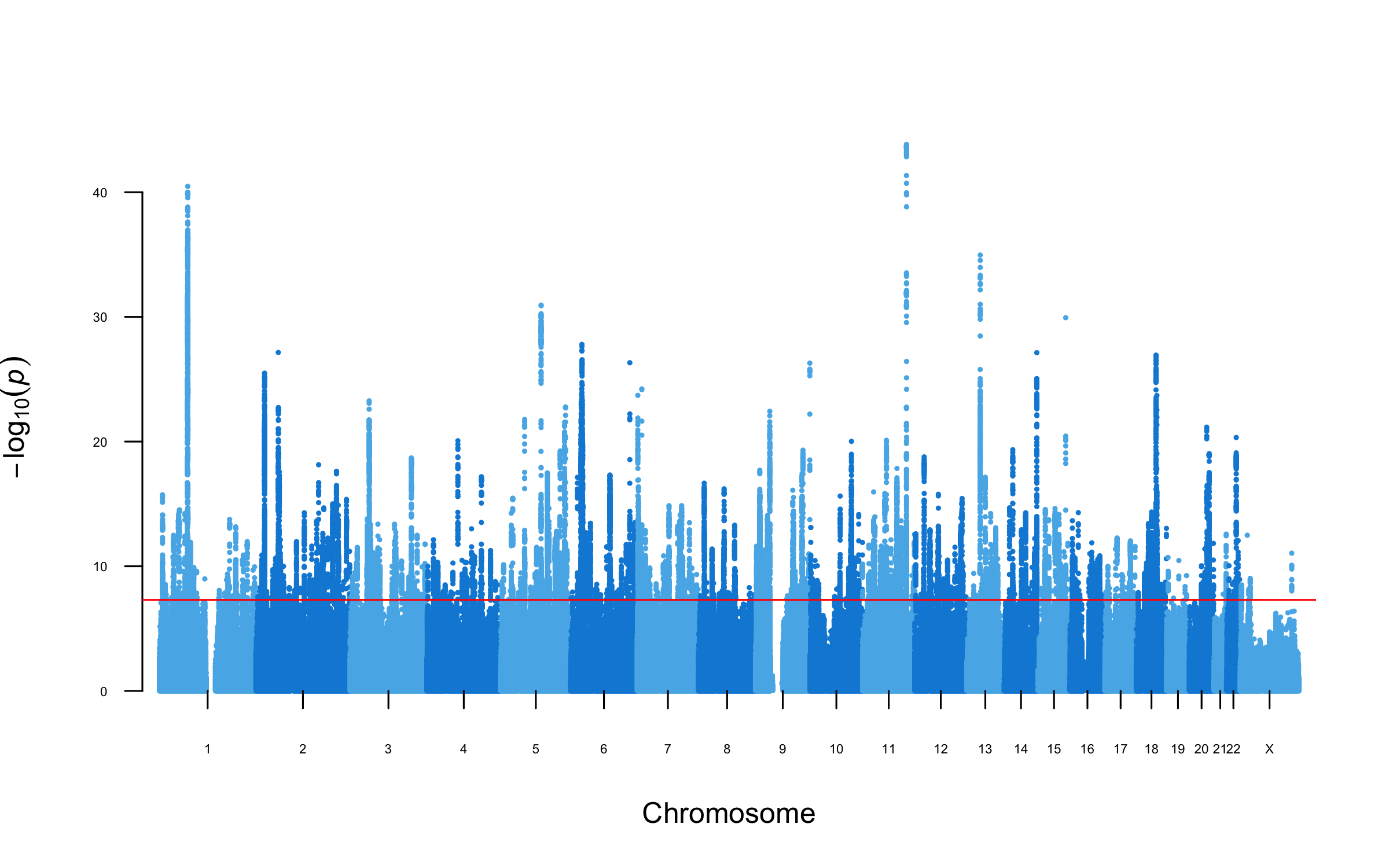
The next major meta-analysis ("MDD3") by the Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. See table of included cohorts.
Current version: v3.49.46.01 [DIV]
Meta-analysis of cohorts for genome-wide association studies of Major Depressive Disorder.
Analysis conducted on LISA.
Clone the repository
git clone git@github.com:psychiatric-genomics-consortium/mdd-wave3-meta.git
cd mdd-meta
You will need to ensure you have generated and added SSH keys between your machine (OS/Windows/Linux) and your github account. This video gives instructions for how to do this.
Install Anaconda.
Linux:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
sh Miniconda3-latest-Linux-x86_64.sh
MacOS:
bin/bash -c "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install.sh)"
brew install anaconda
Install Python 3.8, Snakemake 5.32, and the basic project dependencies
conda activate base
conda install python=3.8
conda install -c conda-forge mamba
mamba install -c bioconda -c conda-forge snakemake-minimal==5.32.2
mamba install dropbox
mamba install pandas
Configure the analysis workflow. Make a copy of the configuration file
cp config.yaml-template config.yaml
Then edit and fill in config.yaml with the required parameters for the analysis to be conducted.
Run the meta analysis for the required population. If you are only doing downstream analysis on meta-analysed sumstats, go to Step 5.
A meta-analysis can be run for each ancestries group. For example, the European ancestries meta analysis can be run with:
snakemake -j1 --use-conda postimp_eur
See more about adding additional cohorts to the meta-analysis. The meta analysis workflow is stored in rules/meta.smk.
Prepare for running downstream analysis.
Download the required summary statistics "MDD3 202X excluding 23andMe (European Ancestries)" from the PGC website. Move the file into the location
results/distribution/daner_pgc_mdd_no23andMe_eur_hg19_v3.TBD.TBD.gz
Depending on the version of conda you have installed on your machine, you may need to use a package called 'pulp'. This can be installed using pip
pip install pulp
Check the rules directory for the analyses to be run. See more on how to contribute.
- Mark James Adams - analyst - Edinburgh
- Swapnil Awasthi - analyst - Broad
- Fabian Strait - analyst - CIMH
- Xiangrui Meng - analyst UCL
- David Howard - analyst - KCL
- Jonathan Coleman - analyst – KCL
- Oliver Pain - analyst - KCL
- Xueyi Shen - analyst - Edinburgh
- Shuyang Yao - analyst – Karolinska
- V Kartik Chundru - analyst Wellcome Sanger
- Karmel Choi - analyst – Harvard/MGH
- Karoline Kuchenbaecker - analytical group lead - UCL
- Naomi Wray - analytical group director - Queensland
- Stephan Ripke - analytical group director - Broad
- Cathryn Lewis - workgroup chair - KCL
- Andrew McIntosh - workgroup chair - Edinburgh
This project is licensed under the MIT License - see the LICENSE.md file for details
The PGC has received funding from the US National Institute of Mental Health (5 U01MH109528-04). Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480-05-003) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam.