A tensorflow Implementation of dagmm: Deep Autoencoding Gaussian Mixture Model for Unsupervised Anomaly Detection, Zong et al, 2018. Furthermore, I built the convolutional autoencoder to analyze the data of images instead of fully-connected autoencoder.
I've trained the model successfully on the below packages:
- Python 3.6.4
- Tensorflow 1.5.0(GPU version)
The auther encode the input to a vector of a very low dimension(only 1 or 2). This is too small since I am doing
the anomaly detection for images with the size 128 x 128. I use a meta dense layer to map the encoded vector into
the into the low dimension space. Therefore, I could have a better reconstruction error and a trainable GMM as well.
- This tf model is used to perform a real case of anomaly detection for my job. I cannot provide the dataset I used due to the commercial security.
- I've done some experiments on using several autoencoders(compressions namely in the paper) to analyze the important regions as experts suggested in each image. The results didn't perform well however.
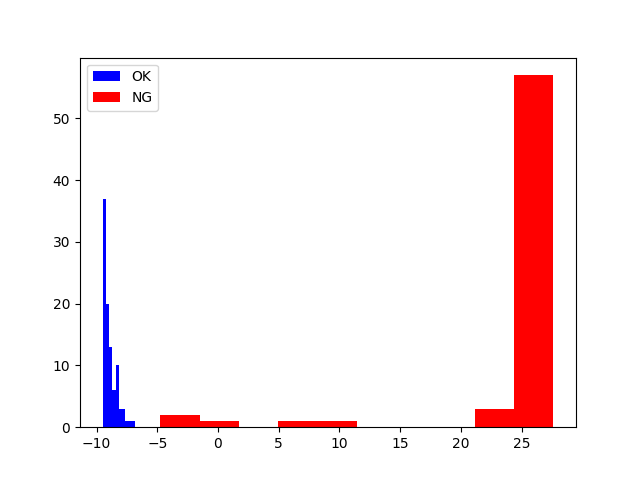
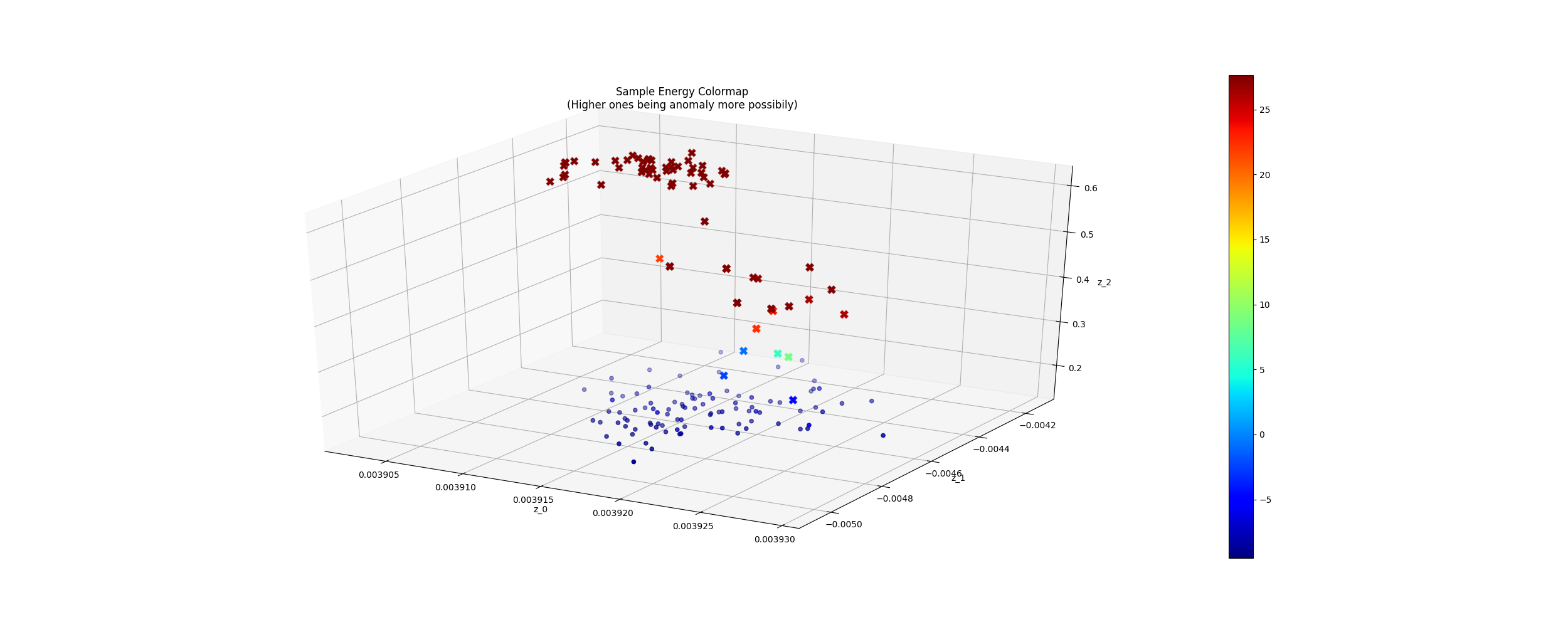
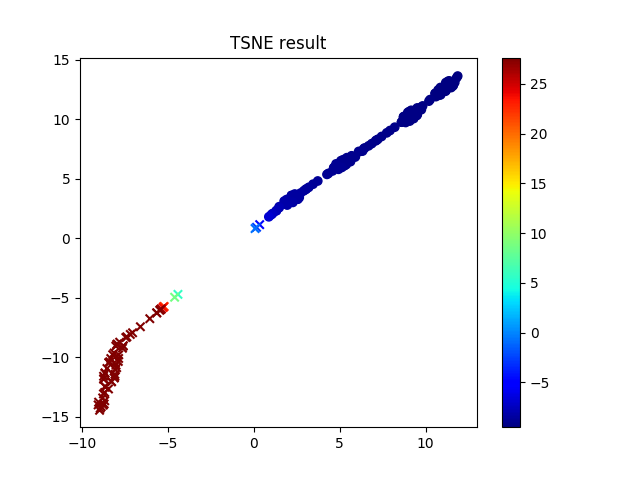
- In my case the required training time is short. The results seem good in less than 2000 epochs
Image configuration needs to be passed through the argument --pattern, a typical setting is like:
OBJECT_J0602 = {
'main': {'width': 128, 'height': 128, 'region': 'all', 'filters': FILTERS_128, 'scope': 'main', 'reuse': False},
}
For example, if we want to use this configuration, --pattern should be argumented with J0601. Some important settings:
- Dictionary key,
'main': The name of interesting region. - width(
int): Specifying the resizing width. - height(
int): Specifying the resizing height. - region(
str,tuple,listoftuple): Region options,'all'means the full image. It could be assigned by a sub-region by a tuple of(xmin, ymin, xmax, ymax)or a list of tuples as well. If it is a list, these sub-regions will assemble the output image vertically. - filters(
listofint): The dimensionality of layers in the encoder. They will be used in the decoder in a reversed direction.
- epoch: The total epochs should be trained.
- encoded_dims: The encoded dimension of the compressor(autoencoder) network.
- latent_dims: The dimension of latent variables of encoded vector.
- lambda1: Hyperparameter tuning in the objective function.
- lambda2: Hyperparameter tuning in the objective function.
- mixtures: The number of mixtures in the gaussian mixture model.
- logdir: The folder for saving the information of this training.
- train_folder: The folder of training data.
- validation_folder: The folder of validation data.
- batch_size: The batch size in each training loop.
Example script:
$ python ./tf-dagmm/train.py --encoded_dims 160 \
--latent 6 \
--mixtures 7 \
--pattern J0601_S \
--logdir /home/i-lun/works/smt/j0601/nsg_split \
--batch_size 56 \
--epoch 1000 \
--train_folder /mnt/storage/P8_SMT/Connector/J0601/wuchi/split/train/OK/ \
--validation_folder /mnt/storage/P8_SMT/Connector/J0601/wuchi/split/test/OK/
Example script:
python ./WiML4AOI_SMT_SA/dagmm/main.py --logdir /home/i-lun/works/kb/type12567hook1 \
--checkpoint checkpoint-1000 \
--saved_in /home/i-lun/works/kb/reports/type12567hook1 \
--train_folder /mnt/storage/AOI_KB/dataset/clean/type12567hook1/train/OK \
--test_OK_folder /mnt/storage/AOI_KB/dataset/clean/type12567hook1/test/OK \
--test_NG_folder /mnt/storage/AOI_KB/dataset/clean/type12567hook1/test/NG
I cannot avoid the singularity issue of gmm, even if the penality term mention in the paper is added. Therefore, I added some jitter to the diagonal of the covariance matrix. Not sure it's a good solution or not.