SVD very very slow and GELS gives nans, -inf
danielhanchen opened this issue · 26 comments
Hey PyTorch devs! First ever issue from me! :)
Anyways, I was investigating the speed of many libraries including Numpy, Numba JIT, PyTorch, Scipy, and figuring out which libraries perform the best on 3 tasks so I could make ML faster for HyperLearn! https://github.com/danielhanchen/hyperlearn/.
- SVD / Pseudoinverses
- Eigh, Eigenvalue decomp
- Lstsq Solve, Least Squares solving.
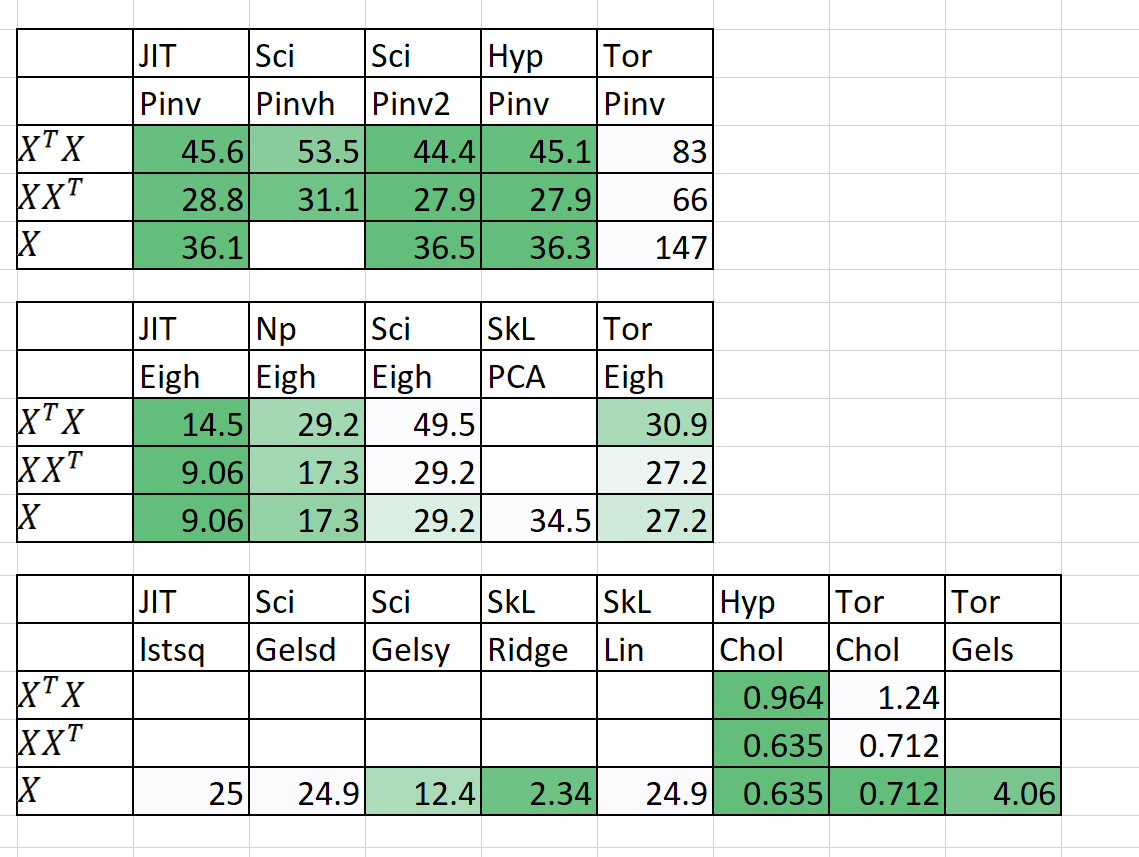
Results in seconds are below given N=5,000 and P=6,000
Now, as you can see for all functions, PyTorch is slower. SVD is really shocking, taking approx 2 or so minutes, whilst Scipy takes approx 30ish seconds. I'm guessing it's because I used the divide and conquer SVD, and assuming that PyTorch is not divide and conquer (hence maybe why I'm seeing not >60% CPU usage --> I set num_threads == max, I'm only seeing 30%)
Also, Eigh is ok. It's very close to Numpy's Eigh, but still a bit suspicious for XXT, where over 10seconds difference is seen.
Finally, Gels is really bad. I do have to disclose I am using a rank deficient matrix (maybe that's why gels is failing?), but there are nans and -infs. I'm assuming division by 0 is causing the error. If you can't fix this, maybe placing theta_hat[ np.isnan(theta_hat) | np.isinf(theta_hat) ] = 0 can solve 1/2 of the problem. However, Gels is surprisingly fast when compared to say Scipy / Numpy's lstsq. Just its super unstable and MSE of the results are really high.
Anyways great work on the package! An extra longer prelim results is @ unslothai/hyperlearn#7
- PyTorch or Caffe2: PyTorch
- How you installed PyTorch (conda, pip, source): conda
- Build command you used (if compiling from source): --
- OS: Windows 10
- PyTorch version: 0.4.1 --> used set_num_threads to max
- Python version: 3.6.6
- CUDA/cuDNN version: -- (using CPU)
- GPU models and configuration: --
- GCC version (if compiling from source): --
- CMake version: --
- Versions of any other relevant libraries: MKL latest (i hope)
cc @jianyuh @nikitaved @pearu @mruberry @heitorschueroff @walterddr @IvanYashchuk @VitalyFedyunin @ngimel
Re SVD: yep we use gesvd (not gesdd). Maybe we should switch.
Thanks for your work and a detailed bug report!
@ssnl Thanks! My first ever bug report so wanted it to be extensive :).
Ohh ok. Would it be possible for pytorch to use gesdd? The divide and conquer would make it approx 2 times or more faster, as it solves SVD by chopping it into 2 parts (divide so can be done in parallel), then combining.
Also, do you know anything about why gels is returning nans, -infs? Thanks!
@ssnl I think there is a faster way to compute Pseudo-inverses, without the usage of SVD. I'll probably look at it.
Re SVD: I just found out that scipy uses gesdd instead of gesvd. Maybe we should really change this. What do you think?
This is a good thread on gesdd vs gesvd: https://groups.google.com/forum/#!msg/julia-dev/mmgO65i6-fA/5OUDc7_nS3wJ
Jack Poulson writes on that thread:
The appropriate reference is LAPACK Working Note 88:
http://www.netlib.org/lapack/lawnspdf/lawn88.pdf
Though it is not explicitly discussed in that paper (AFAIK), the main reason one would choose the implementation based upon the QR algorithm (dgesvd) rather than the one which uses a Divide and Conquer algorithm (dgesdd) is that the latter one requires O(min(m,n)^2) workspace, where k=min(m,n), while the former only requires O(max(m,n)). See the descriptions of LWORK in the following files:
http://www.netlib.org/lapack/double/dgesvd.f
http://www.netlib.org/lapack/double/dgesdd.f
- On the CPU, we should switch to
gesdd. - On the GPU, memory is a premium, so we should prefer
gesvd, which takes up much lesser workspace area.
Could I work on switching CPU SVD from gesvd to gesdd then?
@vishwakftw go for it!
On another note sorry for annoying you guys again, do you know why torch.gels is not performing correctly?
@vishwakftw https://math.stackexchange.com/questions/2076244/find-the-pseudo-inverse-of-the-matrix-a-without-computing-singular-values-of?rq=1
This might interest you.
Also I'm researching on approximate Pseudoinverse computation. I'm using regularized eigenvalue decomp and I'm seeing extremely promising results where EigH Pinv implementation takes just under 3.5seconds for N=1,000,000 P = 200. Whilst Scipy/Numpy's Gesdd takes 12.5 seconds (approx 4x faster).
However, a cost is paid. Numercial stability is sacrificed, with approx 200% more error when compared to exact SVD, where eigH sum(abs(inv @ X)) = 554, whilst SVD's = 217 (so double). Diagonal entries sum to 159.883 and SVD is 159.071 so really really close. Testing on a Least Squares problem, X \ y, EigH pinv's MSE is 35901.710336, whilst SVD is 35908.968448 (so SVD is higher...)
So new EigH solution is approx 4x faster + a bit better accuracy on least squares soln, at the cost of 2x numerical precision errors.
I think it's never really good to sacrifice so much precision for the sake of speed. There is a paper I read whilst implementing the generic version of pinverse, but never got around to implementing that paper.
@danielhanchen This might be of interest to you:
https://research.fb.com/fast-randomized-svd/
https://github.com/facebook/fbpca
@soumith thanks for the links! I'll check them out ! @vishwakftw ye i guess ur right on the errors part.
Re gels: we are using the QR-based gels routine (not gelsd, gelss, gelsx, or gelsy). It really only works when input matrix is full rank (see http://www.netlib.org/lapack/lug/node27.html)
Ohhhh @ssnl no wonder (matrix i put was purposely rank deficient to test robustness). Hmmm ok.
BTW, I was also recently looking at the speed, here's a benchmark with some current numbers for 1534 x 1534 matrix SVD
numpy default min: 341.51, median: 342.65, mean: 408.34
numpy gesvd min: 1264.59, median: 1265.48, mean: 1266.08
numpy gesdd min: 341.22, median: 341.69, mean: 342.37
TF CPU min: 1279.98, median: 1285.51, mean: 1292.32
TF GPU min: 6962.91, median: 7006.48, mean: 8967.89
PyTorch CPU min: 1048.54, median: 1226.51, mean: 1269.30
PyTorch GPU min: 506.14, median: 511.30, mean: 513.09
@ssnl Just like SVD, do you think we should switch over to gelsd for gels for CPU alone?
The subroutine xGELSD is significantly faster than its older counterpart xGELSS, especially for large problems, but may require somewhat more workspace depending on the matrix dimensions.
@vishwakftw I'm not so sure about that, for two reasons:
gels(what we currently use) is actually much faster than bothgelssandgelsdfor the full rank case.- Using
gelsd/s/y/xfor CPU while usinggelsfor CUDA will introduce the inconsistent behavior where CPU code works with rank deficient matrices, but CUDA code doesn't.
What we can do, is to introduce a flag on torch.gels for users to specify whether their matrix might not be full rank, and use gelsd/s/y for CPU & CUDA. (We may want to chose a different one for CUDA vs. CPU due to memory constraints on CUDA)
Also a pure suggestion, since GELS works only on full rank matrices, wouldn't it be faster for PyTorch to use Cholesky Decomp instead of QR / LU? (Since Choleksy has O(1/3n^3) time, much faster than LU by a factor of 2.
@yaroslavvb (not relating exactly to PyTorch, but could impact PyTorch's symeig (EIGH)) I checked out your analysis on reddit, just a note I think I might have fixed Scipy's EIGH's issues. I submitted a request to change Scipy's to SYEVD for EIGH. I think they just did a simple bug...
scipy/scipy#9212
An easy fix I tried is:
scipy.linalg.eigh(XTX, b = np.eye(len(XTX), dtype = X.dtype), turbo = True, check_finite = False)Which performs at most 3-4 times faster than doing this:
scipy.linalg.eigh(XTX, b = None, turbo = True, check_finite = False)@ssnl I guess you are suggesting something like matrix_rank. I think it's doable, could I try it out?
Unfortunately, gels in MAGMA doesn't have many variants unlike LAPACK, which makes me wonder if gels in MAGMA is capable of operating on rank-deficient matrices at all. I was also unable to find any note in MAGMA that suggests the usage of gels for rank deficient matrices.
For SVD, it would be great to offer the user the option of trading memory for speed, I think. Is it possible to dispatch dynamically to gesdd or gesvd?
I got the same issue here…
temp.shape
Out[14]: torch.Size([1140, 1140])
which has low rank structure
u,s,v = torch.svd(temp)
it took 3.5 seconds!
vs
u,s,v = torch.svd(temp.cpu())
it took 0.23 seconds!
ridiculously slow here…
Is there any update on this?
I also ran into this issue when computing svd for a lot of small matrices:
CUDA:
a = tc.randn(16000, 2, 2, device='cuda:0')
%timeit a.svd()Output:
28 s ± 2.58 s per loop (mean ± std. dev. of 7 runs, 1 loop each)
CPU:
a = tc.randn(16000, 2, 2)
%timeit a.svd()Output:
264 ms ± 38 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
I used a GTX 1050 for this comparison, so relatively low end but it shouldn't be 100 times slower on GPU?
May be #42666 will fix these issues.
We moved to cusolver's jacobi methods in the last year. Now the numbers for the tests in #11174 (comment) #11174 (comment) #11174 (comment) look like this:
[[------------------ SVD ------------------]
| cpu | cuda
1 threads: --------------------------------
shape (16000, 2, 2) | 22 | 11.3
shape (1140, 1140) | 330 | 196.5
shape (1534, 1534) | 782 | 454.8
Times are in milliseconds (ms).
Script
import torch
from torch.utils.benchmark import Compare, Timer
def get_timer(device, shape):
A = torch.randn(shape, device=device)
timer = Timer(
r"""
torch.linalg.svd(A, full_matrices=False)""",
globals={"A": A},
label="SVD",
description=f"{device}",
sub_label=f"shape {shape}",
num_threads=1,
)
return timer.blocked_autorange(min_run_time=5)
def get_params():
shapes = ((16000, 2, 2),
(1140, 1140),
(1534, 1534))
for device in ("cpu", "cuda"):
for shape in shapes:
yield device, shape
compare = Compare([get_timer(*params) for params in get_params()])
compare.trim_significant_figures()
compare.print()So GPU (on an RTX2060) is about x2 faster than CPU, so it looks like this is not an issue anymore.
We also added a gesvd as a fallback algorithm for whenever the jacobi method does not work, so now it should always converge (modulo the usual caveats such as "computing the SVD when the eigengap is small is difficult" and so on).
As a side note, we will work this year in bringing more optimised algorithms for very thin/wide matrices and so on.