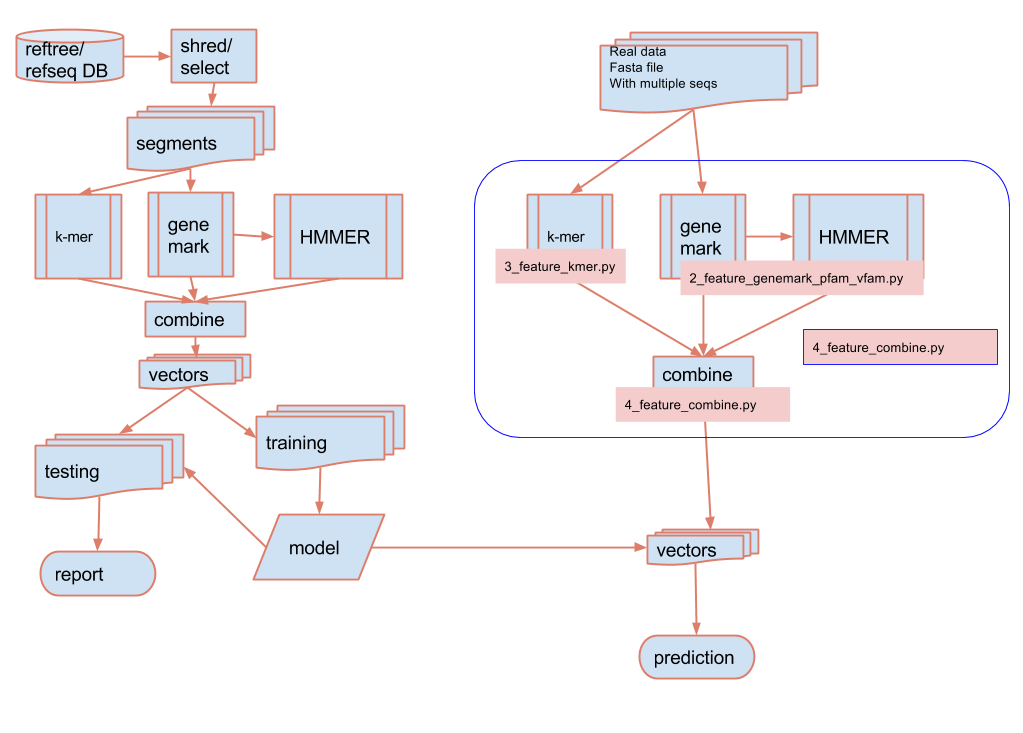
Classifying virus from metagenomic and metatransciptomic contigs
An updated version of ViCA using Deep Learning approach is hosted at: https://github.com/USDA-ARS-GBRU/vica
With this package, a model is offered with training using simulated data from RefSeq genomes.
Tools are provided if the users want to train the model themselves with their own data. Please refer to documentation.
There are three use cases for doing the prediction:
pipeline (in NextFlow) used for prediction on large number of sequences using HPC or Cloud system
scripts/feature_extraction.nf
$SPARK_PATH/bin/spark-submit spark_prediction.py
usage: spark_prediction.py [-h] libsvm model scaler outfile
downloadable package used for prediction on small number of sequences running locally (like a laptop)
~/scripts/prediction_pipeline_lite.py
usage: prediction_pipeline_lite.py [-h]
input_file output_file genemark_path
hmmer_path hmmer_db spark_path feature_file
model_directory scaler_directory
a web interface where the users can submit small number of sequences for prediction
~/web/server.py
Please refer to documentation for dependency and installation details