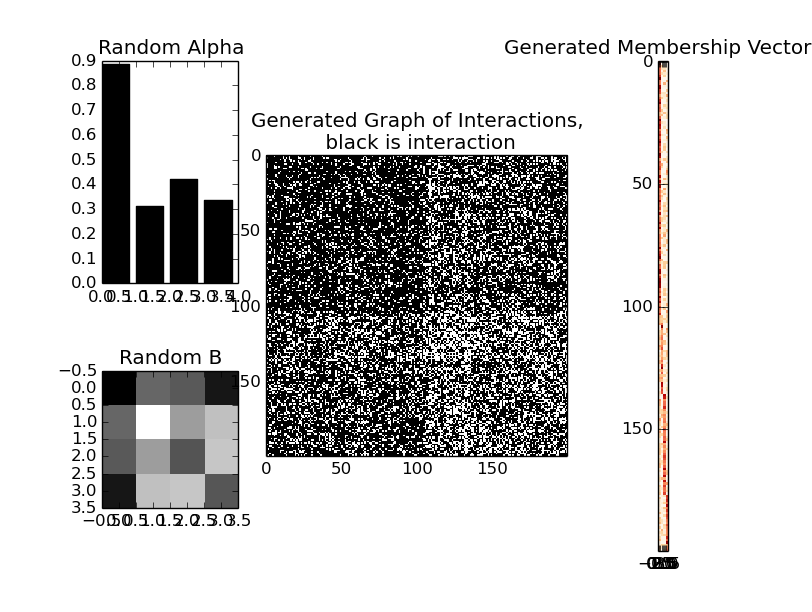
Made for MIT class 6.882 (Spring 2016), based on Mixed Membership Stochastic Blockmodels by Edoardo M. Airoldi, David M. Blei, Stephen E. Fienberg and Eric P. Xing
- with visualizations for alpha, B, pi, sampled G
- randomized alpha (Dirichlet paramater) and interaction matrix between groups
- automatically sorts each variable by argmax cluster membership vectors (similar to what has been done in the paper)
- includes sparsity parameter, rho
python mmsb.py -K [whatever K you want] -N [some number of items] -a [alpha](optional, if not entered it will be randomized) -r [rho] (optional sparsity paramter)
or see help using python mmsb.py -h