Code to visualise mitotic timing and progression of mitosis using IGOR Pro.
Summary statistics including cumulative histograms and stick graphs are plotted from data acquired by the user.
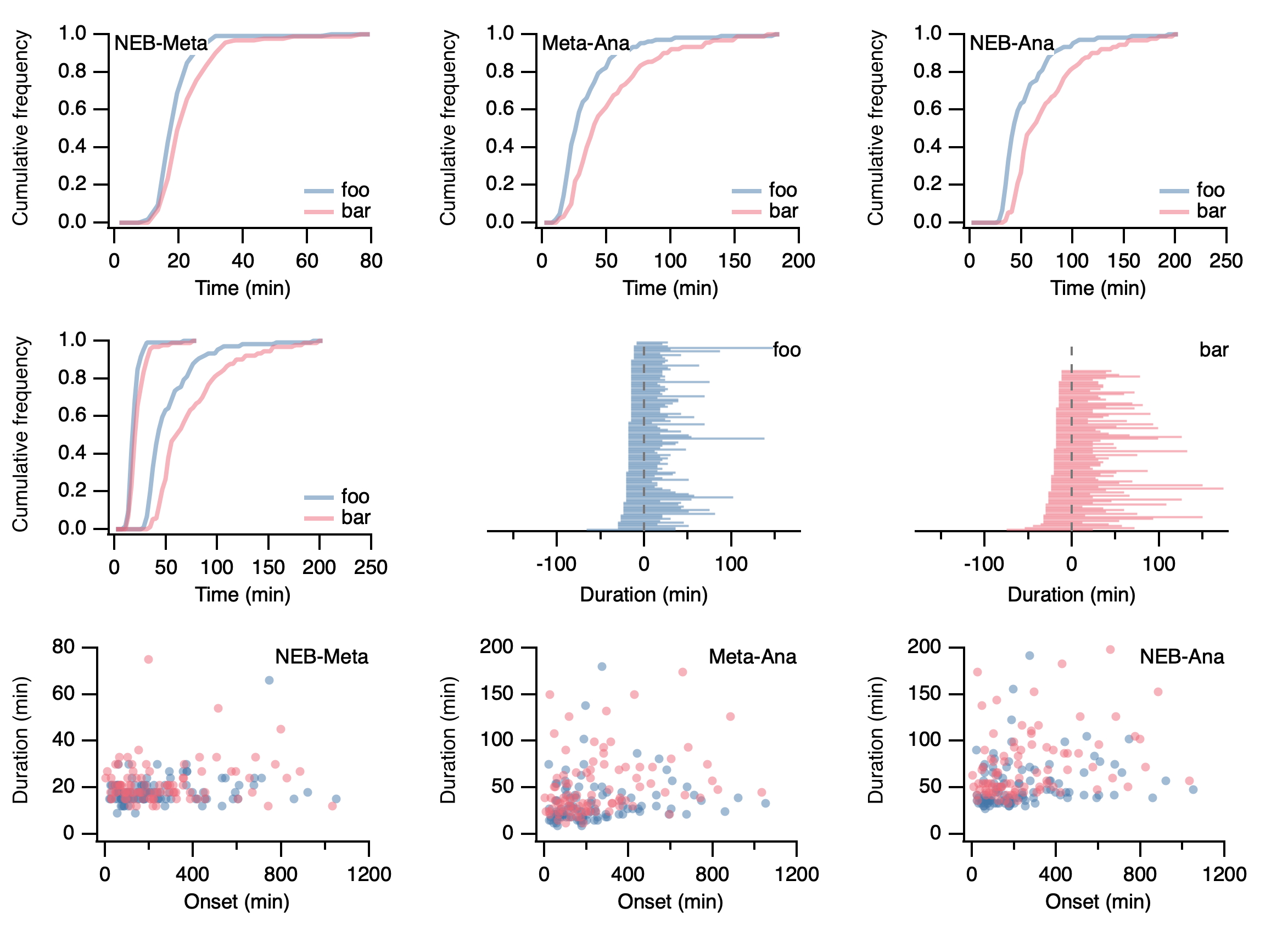
An example file is included in the repo to try. A typical output is shown below.
In addition, the following summary is printed:
foo_NM progression: 106 for 106 cells.T-half: 17.357
bar_NM progression: 90 for 90 cells.T-half: 19.7
foo_MA progression: 106 for 106 cells.T-half: 25.8
bar_MA progression: 90 for 90 cells.T-half: 39.5
foo_NA progression: 106 for 106 cells.T-half: 42.375
bar_NA progression: 90 for 90 cells.T-half: 60
User gathers data in an Excel workbook. Each worksheet is a separate condition (label it appropriately) Three columns labelled NEB, Meta, Ana Each row is a cell's timeline, user inputs the frame numbers for transitions.
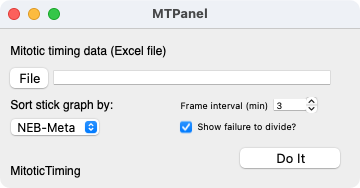
Select Macros > Miotic Timing... in Igor after loading MitoticTiming.ipf
Using this panel, the user specifies the Excel workbook by clicking File.
- The time interval of the experiments can be specified (default is 3 min, if your data is in minutes and not frame numbers, specify 1 here).
- Uncheck the box to remove the cells that do not divide from summary statistics.
- The stick graph can be ordered according to the duration of three different intervals depending on user preference.
MitoticTiming.ipfis written for IGOR Pro 9 but it will compile and run on IGOR Pro 8.- Use NA, NAN or blank to denote cells that didn't transition. Don't use 0.
- It is possible to generate a series of PDF reports from a directory of xlsx workbooks, see Macros menu.