A web app to conduct studies for medical images made using magic of 3d slicer, VNC and web apps.
To proceed with installation docker and docker-compose are required.
Build the docker image from the docker-compose.yml file as follows:
docker-compose build
-
Create a
datadirecotry and move all the DICOM files (.nii.gz files) to thatdatafolder. Also, adddb_nii.csvfile todatadirectory with UID and DICOM file path for each data file inside thedatadirectory.A sample of the
db_nii.csvfile should look likeuid,filepath 1.2.3.4.5,/root/data/1.2.3.4.5.nii.gzWe mount this
datavolume to thedatadirectory inside the docker container. -
Create and run a docker container based on the docker image we build.
Run with logs.
docker-compose up.Run in the detached mode.
docker-compose up -d -
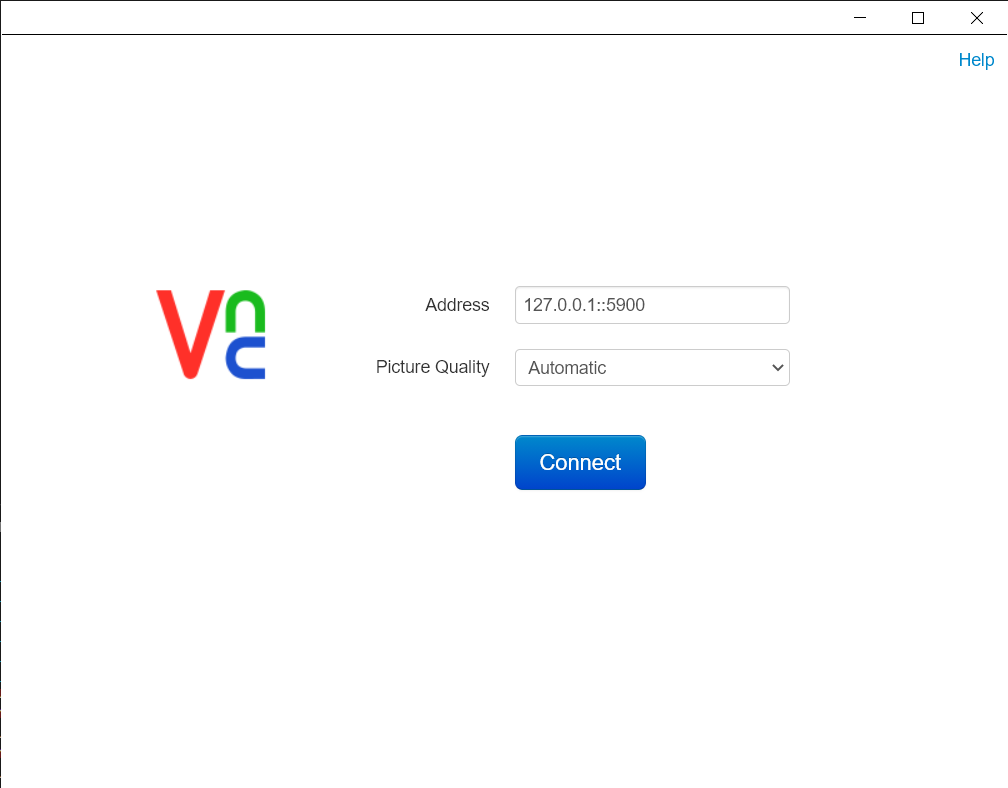
Once docker container is up and running, 3d slicer can be accessed via VNC viewer at
127.0.0.1:5900.Note: The default password for VNC viewer is
vncpassword@123which can be configured indocker-compose.ymlbefore building a docker image. -
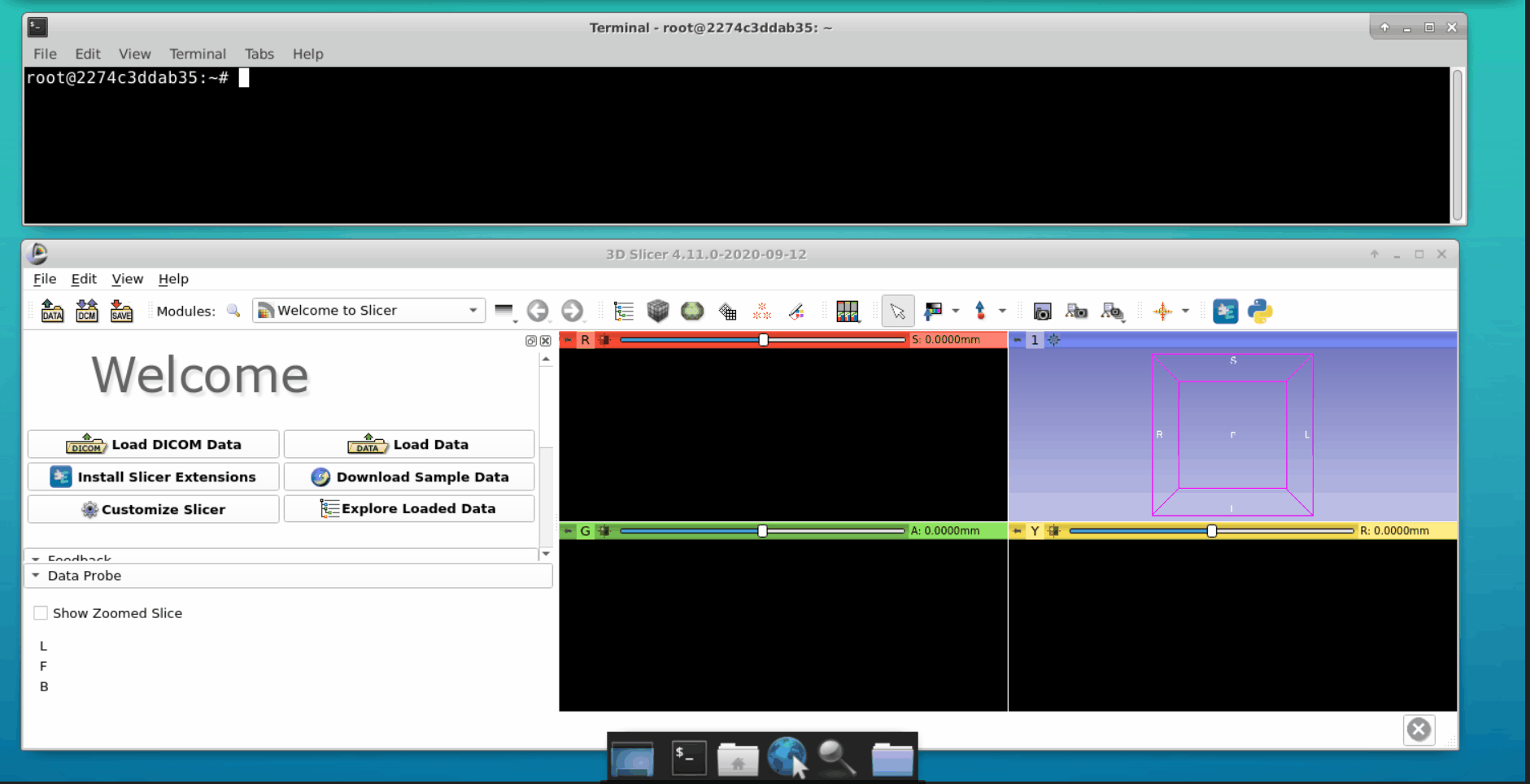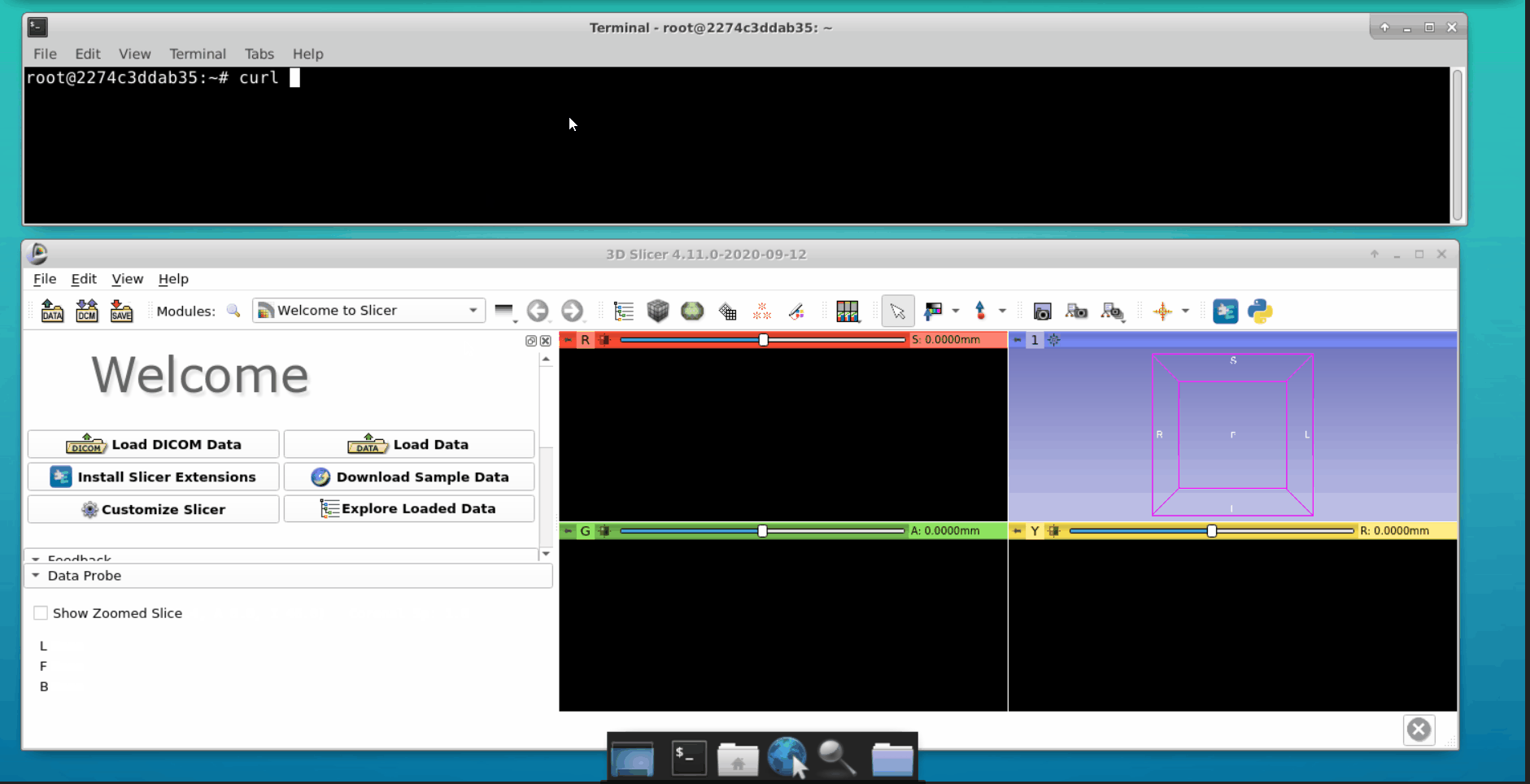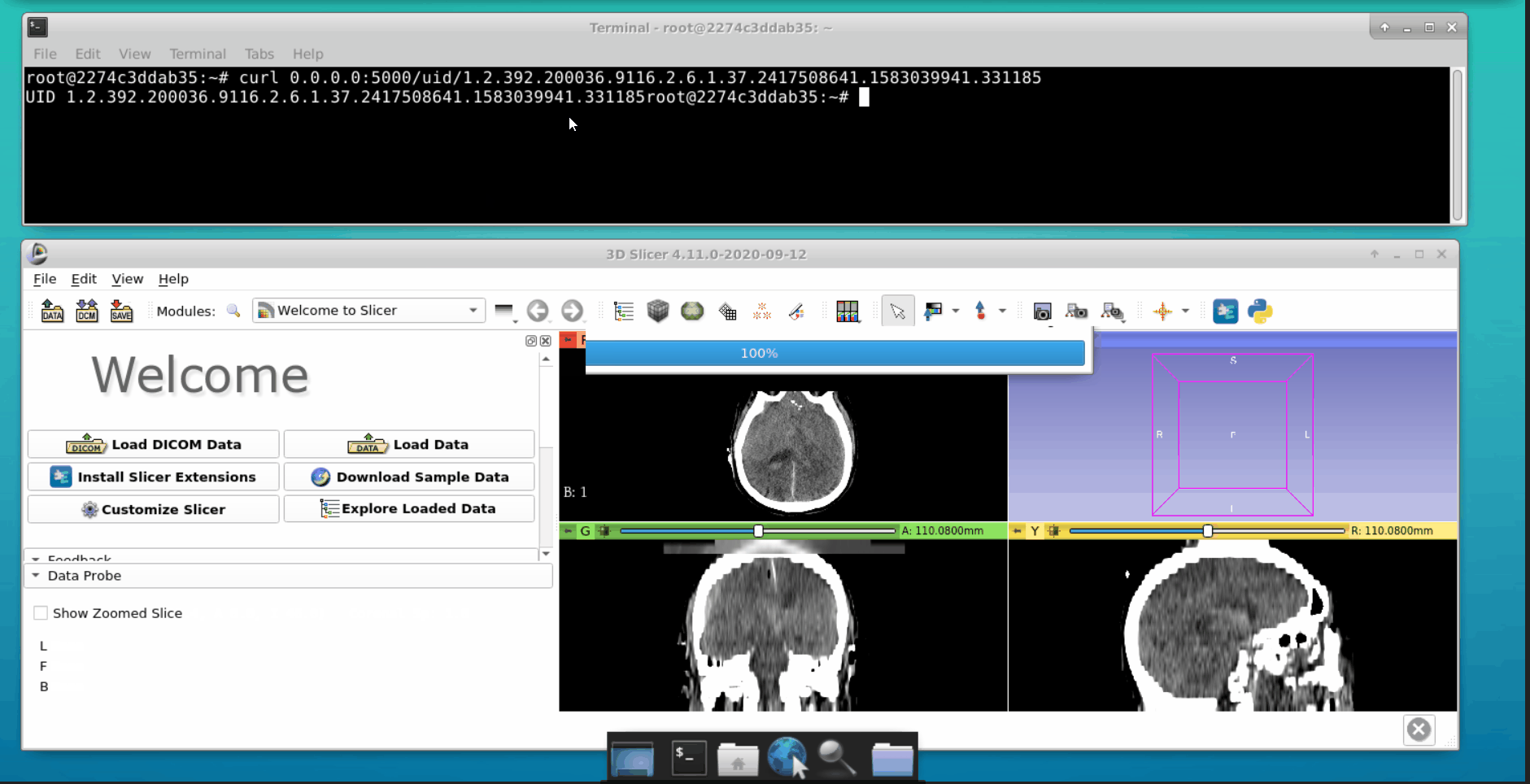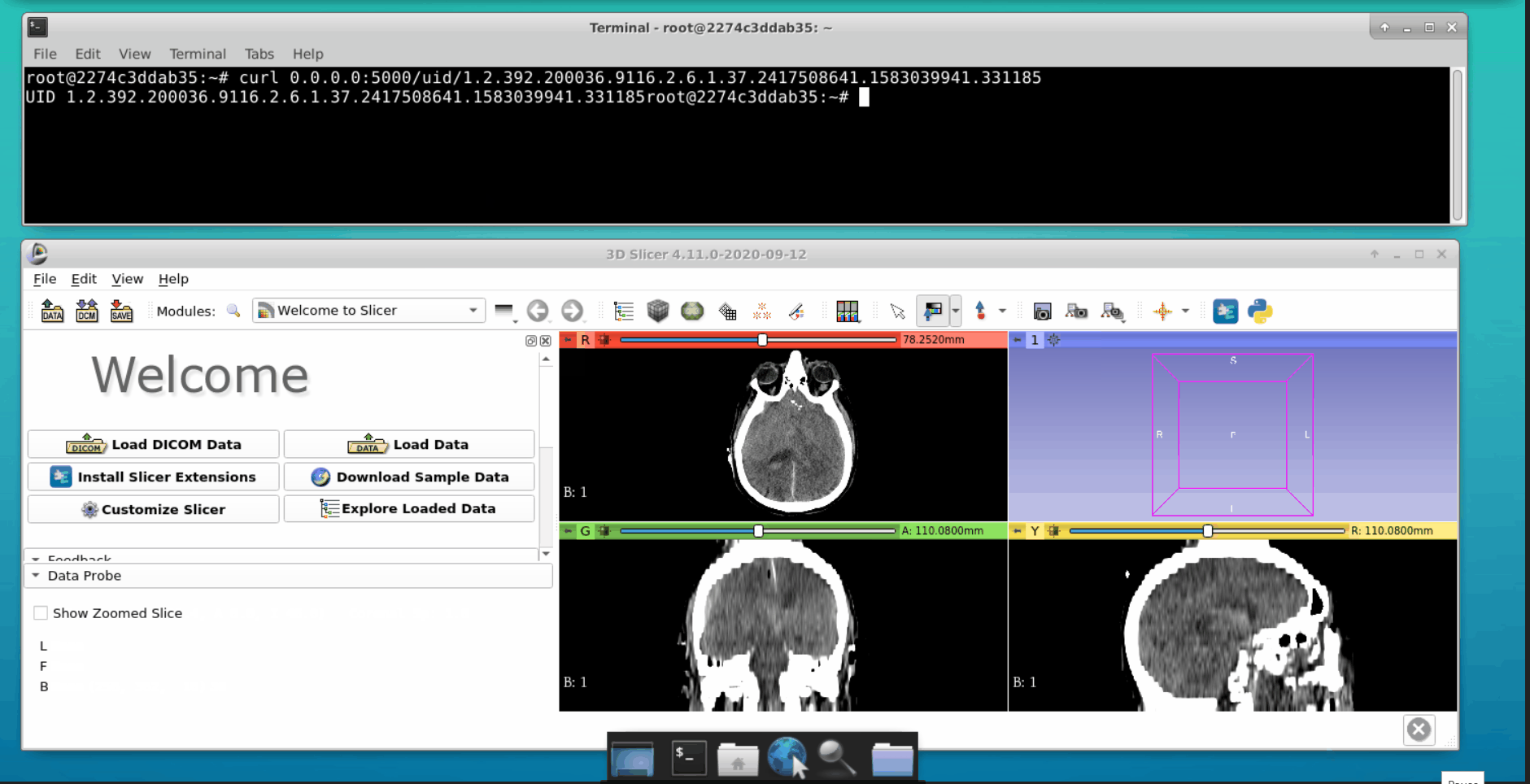
Open a new tab in the terminal and interact with the Flask API. We can load data into the 3d slicer tool as follows:
Note: In case of a failure or issues faced, perform
docker-compose downanddocker-compose up.