The official nnUNet benchmarks for The University of California San Francisco Adult Longitudinal Post-Treatment Diffuse Glioma (UCSF-ALPTDG) dataset.
Please install nnUNetV1 in developer mode.
Please look at the notebooks folder to conduct your own experminets using nnUNetv1. Using that code you can also reproduce the same model files that we are sharing in this paper. The notebook helps to perform standard nnUNetv1 experiments on the dataset.
All important models are available here for download.
These are nnUNetv1 models that have been exported to zip files using the nnUNet_export_model_to_zip command. To use them, all you have run is nnUNet_install_pretrained_model_from_zip, and you will be able to run any of the models.
We provide 3 models for the UCSF-ALPTDG dataset as benchmarks.
- UCSF_PostTreatmentGliomaSegModel: Model that takes inputs as (1) FLAIR (2) T1 (3) T2 and (4) T1CE MR images.
['flair','t1','t2','t1ce'] - UCSF_FLAIRChangeSegModel: Model that takes input as the (1) FLAIR image in Timepoint 1 (2) FLAIR image in Timepoint 2 and (3) FLAIR subtraction image (t2-t1).
['time1_flair','time2_flair','flair_subtraction'] - UCSF_T1PostChangeSegModel: Model that takes input as the (1) T1CE image in Timepoint 1 (2) T1CE image in Timepoint 2 and (3) T1CE subtraction image (t2-t1).
['time1_t1ce-t1','time2_t1ce-t1','t1ce_subtraction']
We also provide 2 skull stripping models for FLAIR and T1Post skullstripping respectively.
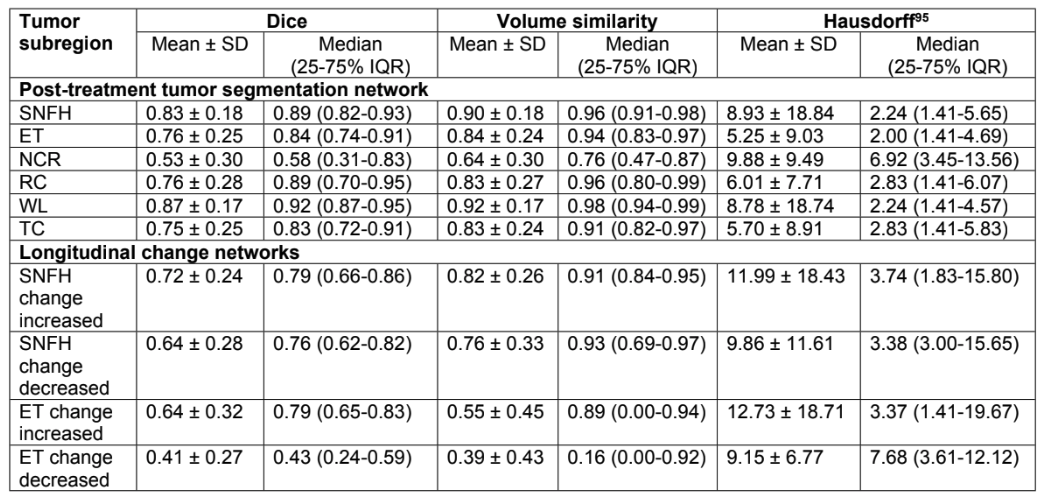
Following are the performance metrics for all the experiments performed in this project.
If you have used this work in your project, considering citing this research paper.
@article{rudie2022longitudinal,
title={Longitudinal Assessment of Posttreatment Diffuse Glioma Tissue Volumes with Three-dimensional Convolutional Neural Networks},
author={Rudie, Jeffrey D and Calabrese, Evan and Saluja, Rachit and Weiss, David and Colby, John B and Cha, Soonmee and Hess, Christopher P and Rauschecker, Andreas M and Sugrue, Leo P and Villanueva-Meyer, Javier E},
journal={Radiology: Artificial Intelligence},
volume={4},
number={5},
pages={e210243},
year={2022},
publisher={Radiological Society of North America}
}
Data and models cannot be used for commerical use.