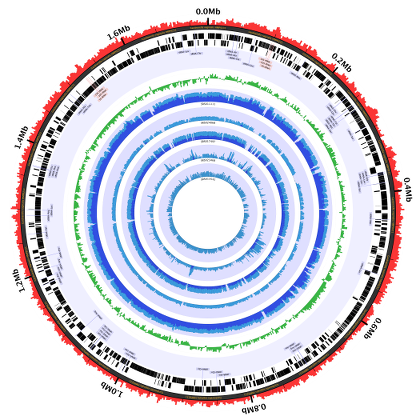
The Charm City Circleator--or Circleator for short--is a Perl-based visualization tool developed at the Institute for Genome Sciences in the University of Maryland's School of Medicine. Circleator produces circular plots of genome-associated data, like this one:
Common uses of the tool include:
- Displaying the sequence and/or genes in a GenBank flat file.
- Highlighting differences and/or similarities in gene content between related organisms.
- Comparing SNPs and indels between closely-related strains or serovars.
- Comparing gene expression values across multiple samples or timepoints.
- Visualizing coverage plots of RNA-Seq read alignments.
Circleator...
- Builds on BioPerl and the input file formats that it supports, including:
- GenBank flat files, GFF, FASTA
- Accepts a number of other commonly-used datatypes and file formats:
- Outputs publication-ready figures in the SVG (Scalable Vector Graphics) format.
- Requires only a single configuration file whose layout mirrors that of the figure itself.
- Predefined configuration files and "track" types are supplied for common datasets.
- Advanced features allow limited analyses to be performed as a figure is drawn.
- Includes an extensive set of regression tests.
- Offers a prototype web-based GUI (under the "Ringmaster" project.)
- Perl 5.6 or later and the following Perl modules/packages:
- BioPerl
- JSON
- Log::Log4perl
- SVG
- Text::CSV
- Vcf
- The Apache Batik package to convert SVG to PDF, JPEG, or PNG.
- The samtools package in order to read SAM/BAM files.
- The vcftools package (which includes the Vcf Perl module) in order to read VCF files.
First, install Circleator. After installing Circleator and its prerequisites, running the program requires as little as:
- A GenBank flat file for a genome of interest.
- A Circleator configuration file.
Examples of both of these types of files can be found in the Circleator source distribution. For example, from the top level of the unpacked Circleator zip or tar file the following command will create a Circleator figure for CM000961.gbk, which contains the genome of Corynebacterium genitalium ATCC 33030:
circleator --data=data/CM000961.gbk --config=conf/genes-percent-GCskew-1.cfg > fig1.svg
The resulting SVG file, fig1.svg can be viewed directly in many
recent web browsers or image viewers. Or, if the Apache Batik
package has been installed, it can be used to convert the image to
PDF, PNG, or JPEG, using a wrapper script (rasterize-svg) from the
Circleator distribution:
rasterize-svg fig1.svg pdf
rasterize-svg fig1.svg png
rasterize-svg fig1.svg jpeg
The Circleator configuration file format aims to make the common case fast and the uncommon case possible. In other words, new users should be able to quickly produce a standard visualization of their data, provided it is in a commonly-used format. Conversely, experienced users should be able to create intricately-customized figures by using the same configuration file syntax.
- Walkthroughs and sample configuration files provide HOW-TO guides for commonly-encountered datasets.
- Predefined track types render standard data types using reasonable default options.
- e.g., the keyword
genesby itself on a line in the configuration file will display a circular track in which each gene is rendered as a curved black rectangle.
- e.g., the keyword
- Configurable track options allow the predefined track types to be customized as little or as much as needed.
- e.g., the line
genes color1=redwill behave the same asgenes, but using red instead of black.
- e.g., the line
- User-defined track types can be created inline and then reused later in the configuration file.
- Feature-based scaling allows the figure scale to be selectively expanded around features of interest.
- e.g., Use 100X scale for any nonsynonymous SNP position and use the additional space to display the affected base.
- Configuration file loops mean that figures for 60-genome SNP panels can be configured and displayed without having to cut and paste the same SNP track configuration 59 times.
- Symbolic track references allow tracks to reference others by name or relative position.
- e.g., Label each tRNA displayed in the preceding track with its anticodon sequence and connect the label to the corresponding feature with a blue line.
- Computed features can be added to supplement those features that appear directly in the input files.
- e.g., Create and display a "SNP desert" feature in any location where there is at least 5kb of sequence that contains no SNPs.
- e.g., Create and display a "low coverage" feature in any location where the value plotted in the read coverage graph in track 2 falls below 5.
Circleator is Copyright (C) 2010-2017, Jonathan Crabtree <jonathancrabtree@gmail.com>
Circleator is free software and is distributed under the terms of the Artistic License 2.0. For details, see the full text of the license in the file LICENSE in the top-level of this distribution.
Files with different licenses or copyright holders:
Copyright (c) 2002 Cynthia Brewer, Mark Harrower, and The Pennsylvania State University. The color specifications in this file are covered by an Apache-Style license. Please see the license statement in the file for details. The file itself was prepared by Martin Krzywinski and downloaded from http://mkweb.bcgsc.ca/brewer/swatches/brewer.txt
This product includes color specifications and designs developed by Cynthia Brewer (http://colorbrewer.org).
An Applications Note describing Circleator has been published in Bioinformatics:
Crabtree, J., Agrawal, S., Mahurkar, A., Myers, G.S., Rasko, D.A., White, O. (2014) Circleator: flexible circular visualization of genome-associated data with BioPerl and SVG. Bioinformatics, 10.1093/bioinformatics/btu505.