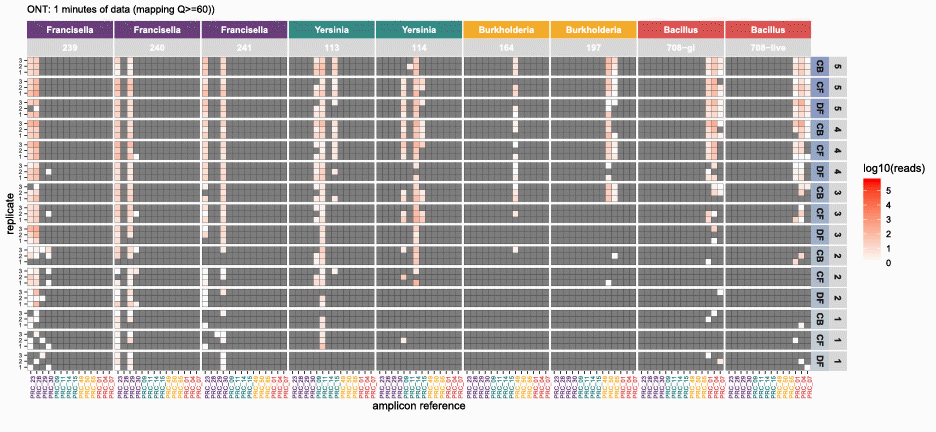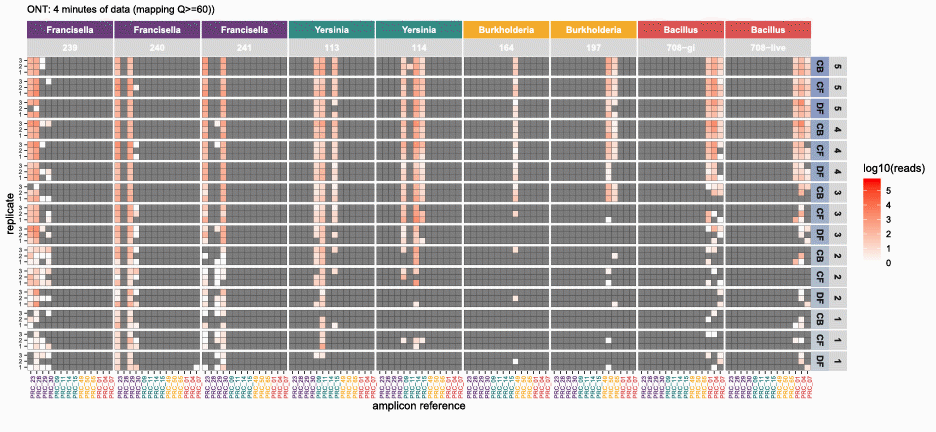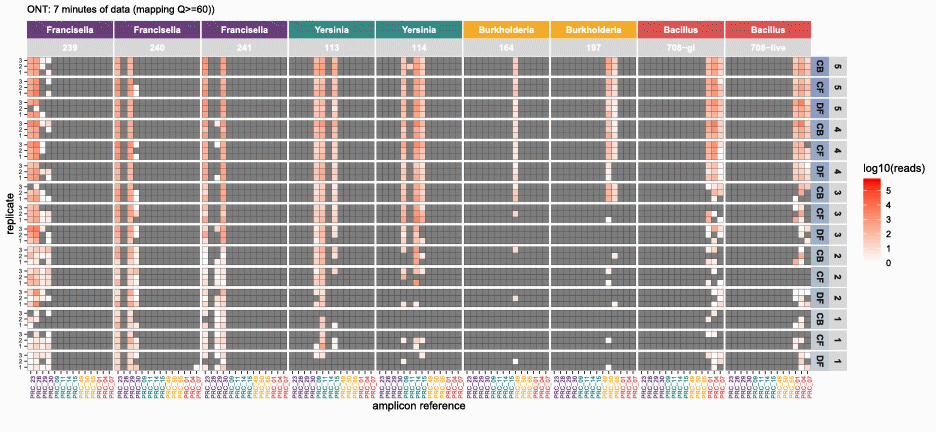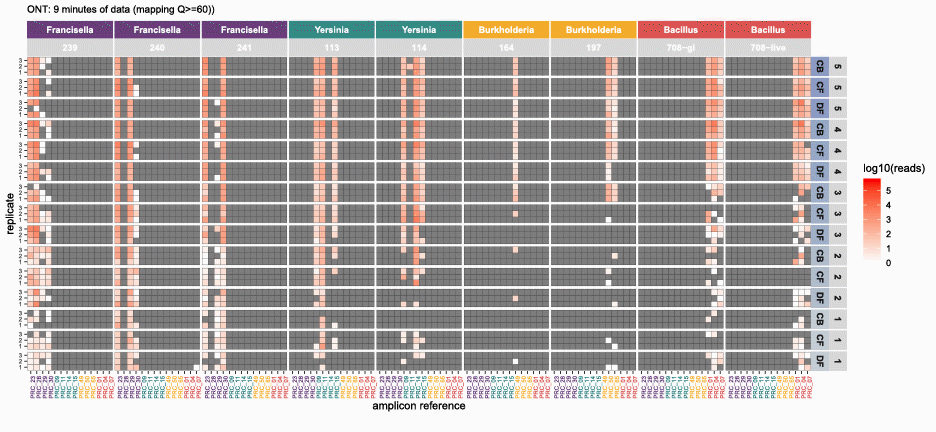
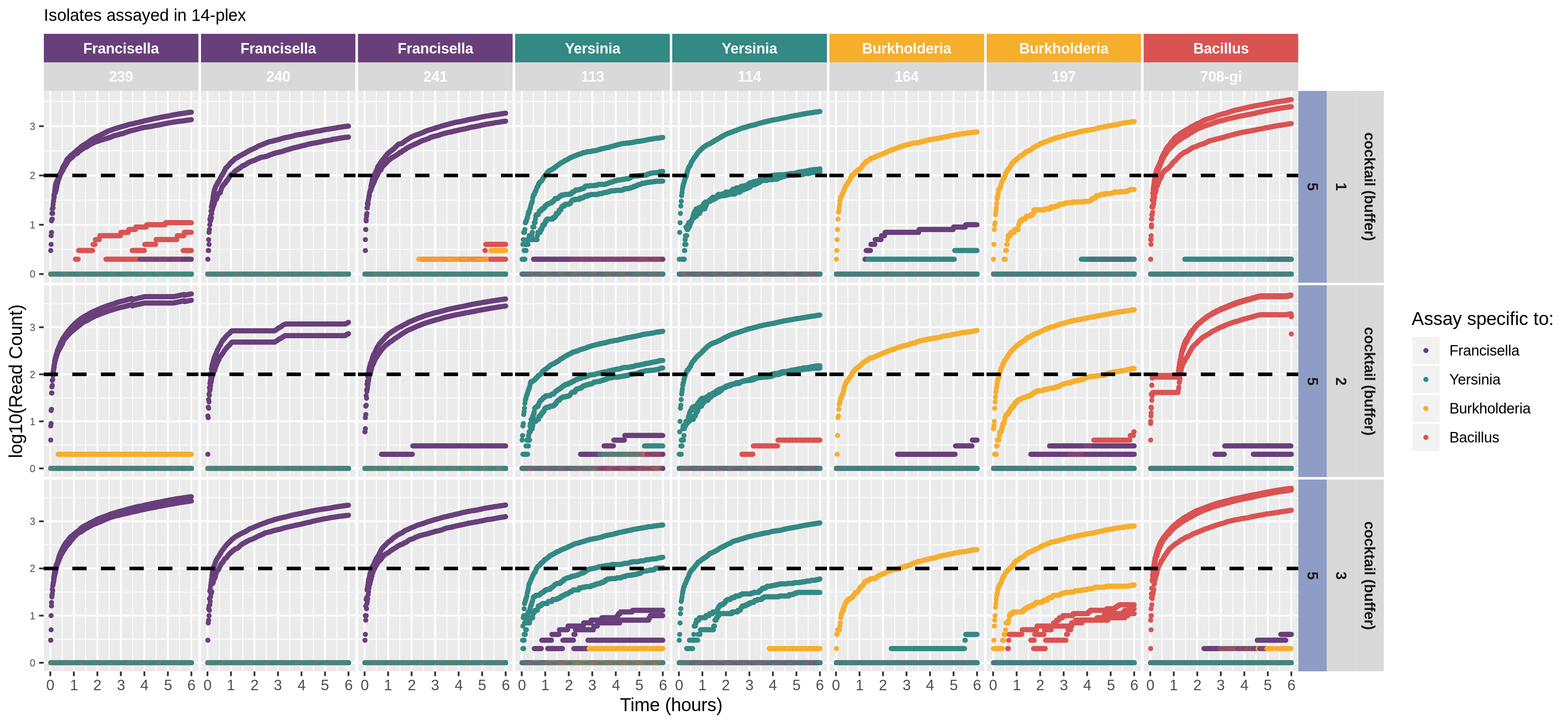
nanotimeparse parses an Oxford Nanopore fastq file on read sequencing start times found in the fastq headers.
It does not require the sequencing_summary_<runID>.txt file output from the MinKNOW software.
It can generate the read data that underpins visualizations like these from (also please cite this publication if using nanotimeparse):
Get subsets of (-i) Oxford Nanopore Technologies (ONT) basecalled fastq reads in slices of (-s) minutes, over a period of (-p) minutes. Input fastq file is output as 2 sets of n fastq files (n = p/s). Set 1 is n fastq files, and each file contains reads generated from the start of the ONT run to each time slice. Set 2 is also n fastq files, but each file contains only newly generated reads between each time slice.
- for best results, p should be evenly divisible by s (i.e. p/s = INT)
- using INT for minutes is preferrable, however, FLOAT is fine if p/s = INT
- example output, see `sandbox/nanotimeparse-i_test.fq.s_1.p_10/`, and check log for details
- be sure to concatenate all fastq files output from a single ONT flowcell
nanotimeparse.sh -t <threads> -i </absolute/path/to/nanopore_basecalled.fastq> -s <minutes> -p <minutes>
-h help help message
-t INT number of threads to GNU parallel over
-i FASTQ input ONT basecalled fastq
-s INT or FLOAT time slice in (minutes)
-p INT or FLOAT period of time to slice up since start of sequencing run (minutes)
Executing with 10 threads (@2.1GHz), and slicing on every hour over a 48 hour period (96 fastq files as output), nanotimeparse takes about 30 minutes to parse 1.2M reads (~15GB) into both sets (and memory maxes out at (15GB/2)*10=75GB, see below). Of course this all depends on read N50 with respect to file sizes and memory requirements.
If memory is limited, use less threads. When running nanotimeparse, each thread requires approximately the size of your input fastq file in memory. Unfortunately this limits the user to a maximum file size equal to the amount of memory on their computer.
For Example:
nanotimeparse.sh -t 10 -i sample.fastq -s 1 -p 5
If input (-i) sample.fastq is 1.0 GB, each thread requires ~1 GB memory. Since we're running with 10 threads, the entire run will require ~10 GB memory to complete successfully.
GNU Parallel [1] and GNU CoreUtils: cat, mkdir, sed, sort, cut, date, paste, basename, printf, comm, grep, awk, uniq, dirname, split, find, head, bc, seq, tail, wc
The only non-GNU CoreUtils dependency is GNU Parallel [2].
If you'd like to call the tool globally, symbolically link the shell script into a $PATH path. For example:
sudo ln -s $PWD/nanotimeparse/nanotimeparse.sh /usr/local/bin
sudo apt install parallel
git clone https://github.com/raplayer/nanotimeparse.git
sudo apt install parallel
brew install coreutils
brew install gnu-sed
git clone https://github.com/raplayer/nanotimeparse.git
Copyright (c) 2020 The Johns Hopkins University Applied Physics Laboratory
This program is free software: you can redistribute it and/or modify it under the terms of the GNU Affero General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU Affero General Public License for more details.
You should have received a copy of the GNU Affero General Public License along with this program. If not, see https://www.gnu.org/licenses/.
- Player, R., Verratti, K., Staab, A. et al. Comparison of the performance of an amplicon sequencing assay based on Oxford Nanopore technology to real-time PCR assays for detecting bacterial biodefense pathogens. BMC Genomics 21, 166 (2020). https://doi.org/10.1186/s12864-020-6557-5
- O. Tange (2011): GNU Parallel - The Command-Line Power Tool, ;login: The USENIX Magazine, February 2011:42-47.