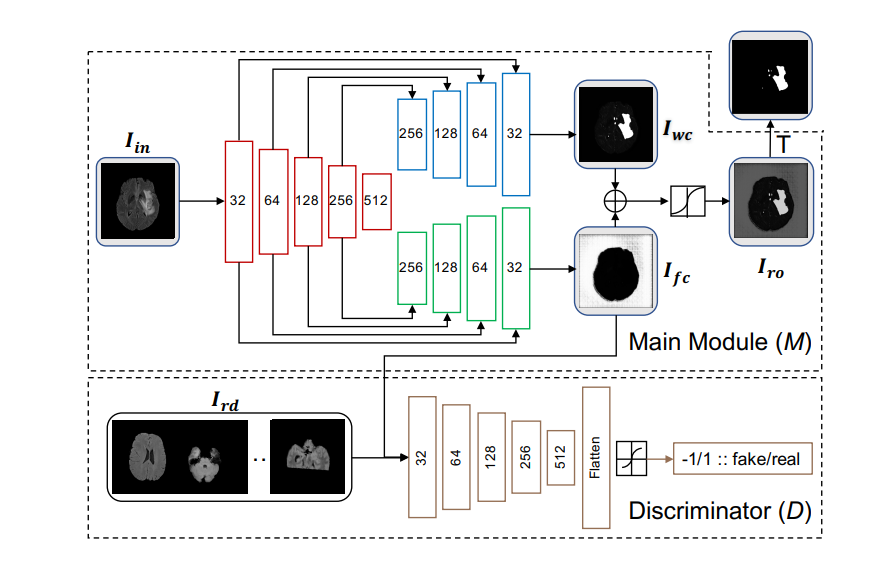
ASC-Net is a framework which allows us to define a Reference Distribution Set and then take in any Input Image and compare with the Reference Distribution and throw out anomalies present in the Input Image. The kind of cases where this is useful is when you have some images/signals where you are aware of its contents and then you get a set of new images and you want to see if the new images differ from the original set aka anomaly/novelty detection.
https://arxiv.org/pdf/2103.03664.pdf
- Solves the difficulty in defining a class/set of things deterministically down to the nitty gritty details. The Reference Distribution can work on any combination of image set and abstract out the manifold encompassing them.
- No need of perfect reconstruction. We care about the anomaly not the reconstruction unlike other existing algorithms. State of the art performance!
- We can potentially define any manifold using Reference Distribution and then compare any incoming input image to it.
- Works on any image sizes. Simply adjust the size of the encoder/decoder sets to match your input size and hardware capacity.
- The claim of "independent of instability of GANs" holds since the final termination is not dependent on the adversarial training. We terminate when the I(ro) output has split into distinct peaks.
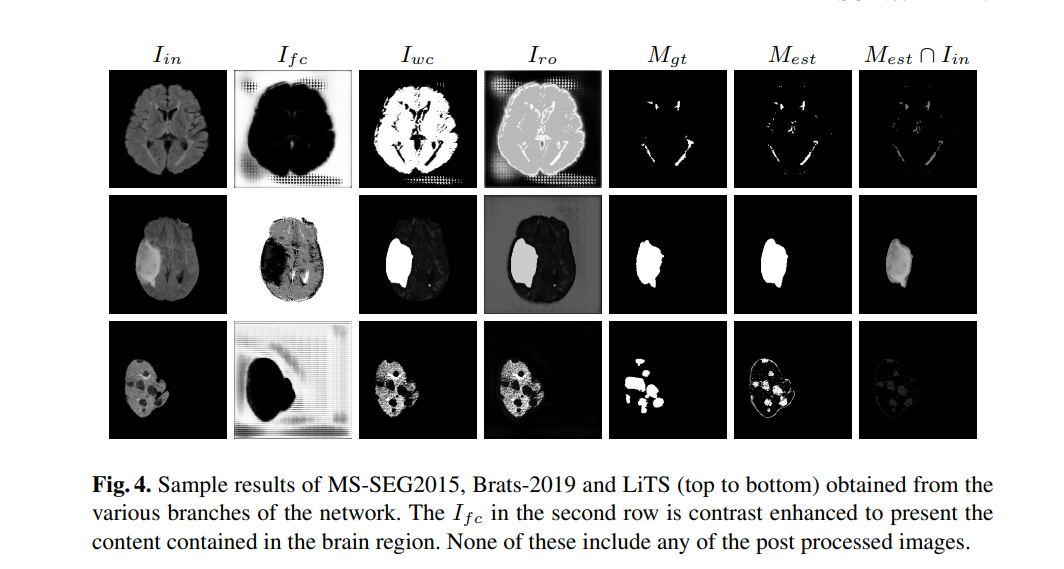
Always take threshold on the reconstruction i.e. ID3 in the code section as it summarizes the two cuts in one place
- CUDA - CUDA-9.0.176
- CUDNN - CUDNN- Major 6; Minor 0; PatchLevel 21
- Python - Version 2.7.12
- Tensorflow - Version 1.10.0
- Keras - Version 2.2.2
- Numpy - Version 1.15.5
- Nibabel - Version 2.2.0
- Open-CV - Version 2.4.9.1
- Brats 2019 - Select Brats 2019
- LiTS - LiTS Website
- MS-SEG 2015 - MS-SEG2015 website
- [12 gb TitanX]
- ID1 is Ifc
- ID2 is Iwc
- ID3 is Iro. Please take threshold on this
-
To make the frame work function we require 2 files [Mandatory!!!!]
- Reference Distribution - Named good_dic_to_train_disc.npy for our code
This is the image set which we know something about. This forms a manifold.
- Input Images - Named input_for_generator.npy for our code
These can contain any thing the framework will split it into two halves with one halves consisting of components of the input image in the manifold of the Reference distribution and the other being everything else/anomaly.
-
Ground truth for the anomaly we want to test for [Optional used during testing]
- Masks - Named tumor_mask_for_generator.npy for our code
The framework is able to throw out anomaly without needing any guidance from a ground truth. However to check performance we may want to include a mask for anomalies of the input image set we use above. In real life scenarios we wont have these and we dont need these.
- The framework is initialized with input shape 160x160x1 for MS-SEG experiments. Please update this according to your needs.
- Update the path variables for the folders in case you want to visualize the network output while training it
- To change the base network please change the build_generator and build_discriminator methods
-
create_networks.py
This creates the network mentioned in our paper. If you need a network with different architecture please edit this file accordingly and update the baseline structures of the encoder/decoder. Try to keep the final connections intact.
- After running this you will obtain three h5 files
- disjoint_un_sup_mse_generator.h5 : This is the main module in the network diagram above
- disjoint_un_sup_mse_discriminator.h5 : This is the discriminator in the network diagram above
- disjoint_un_sup_mse_complete_gans.h5 : This is a completed version of the entire network diagram
- After running this you will obtain three h5 files
-
continue_training_stage_1.py
Stage 1 training. Read the paper!
-
continue_training_stage_2.py
Stage 2 training. Read the paper!