cosmos 
Overview
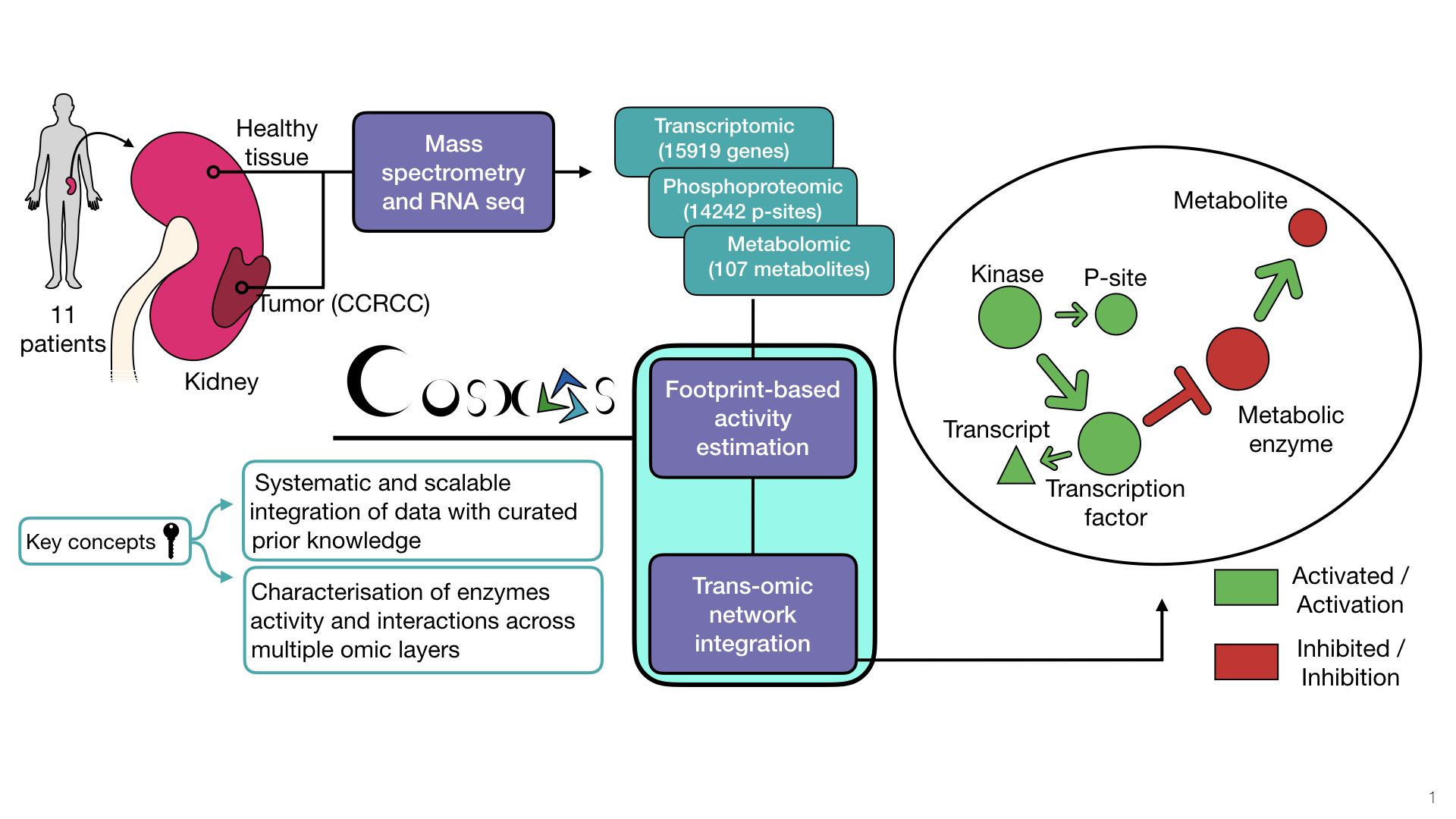
COSMOS (Causal Oriented Search of Multi-Omic Space) is a method that integrates phosphoproteomics, transcriptomics, and metabolomics data sets. COSMOS leverages extensive prior knowledge of signaling pathways, metabolic networks, and gene regulation with computational methods to estimate activities of transcription factors and kinases as well as network-level causal reasoning. This pipeline can provide mechanistic explanations for experimental observations across multiple omic data sets.
COSMOS uses CARNIVAL’s Integer Linear Programming (ILP) optimization strategy to find the smallest coherent subnetwork causally connecting as many deregulated TFs, kinases/phosphatases and metabolites as possible. The subnetwork is extracted from a novel integrated PKN (available here) spanning signaling, transcriptional regulation and metabolism. Transcription factors activities are inferred from gene expression with DoRothEA, a meta resource of TF/target links. Kinase activities are inferred from phosphoproteomic with a kinase/substrate network of Omnipath, a meta resource of protein-protein. CARNIVAL was adapted to find mechanistic hypotheses connecting the TF and kinase activities with metabolites from a signaling/metabolic prior knowledge network combining Omnipath, STITCHdb and Recon3D.
Tutorial
Instal the package (from github with devtools) :
## If needed instal devtool package
install.packages("devtools")
## instal COSMOS
library(devtools)
install_github("saezlab/COSMOS")//!\ Curently avalaible tutorial is available as self contain R script: https://github.com/saezlab/COSMOS/blob/master/tutorial.R A markdown version is in preparation and arriving soon.
//!\ The tutorial.pdf is curently based on the paper version of CARNIVAL, which is the preprint version: https://github.com/saezlab/CARNIVAL/tree/package_paper_version Check the Tutorial.pdf (or click here) file in the root github folder to have an example of running COSMOS with from TF, kinase, phosphatase activities and metabolite abundances. You need to download this release to run the current version of the tutorial. This version is obsolete.
Access
The meta PKN used with the biorXiv version of COSMOS is available here.
An updated meta PKN is available with the package (using load_meta_pkn() in R)
Citation
If you use cosmos for your research please cite the original publication:
Aurelien dugourd, Christoph Kuppe, Marco Sciacovelli, Enio Gjerga, Kristina Bennet Emdal, Dorte Breinholdt Bekker-Jensen, Jennifer Kranz, Eric J.M. Bindels, Sofia Costa, Jesper V Olsen, Christian Frezza, Rafael Kramann, Julio Saez-Rodriguez. Causal integration of multi-omics data with prior knowledge to generate mechanistic hypotheses. bioRxiv 2020.04.23.057893 (2020) doi:10.1101/2020.04.23.057893.
License
The code is distributed under the GNU General Public License v3.0. The meta PKN is distributed under the Attribution-NonCommercial 4.0 International (CC-BY-NC 4.0) License.