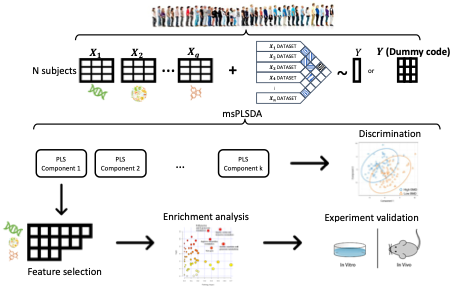
Welcome to the msPLSDA repository!
Multi-omics technologies have revolutionized our understanding of complex biological systems and their association with diseases. These technologies allow us to capture comprehensive molecular information from various biological factors, providing a systems biology perspective. However, integrating and analyzing multi-omics datasets pose significant challenges due to their high dimensionality and heterogeneity. Current methods for multi-omics integration often lack supervised approaches that can extract meaningful insights directly relevant to clinical phenotypes.
In this repository, we present msPLSDA, a novel method for multi-omics data analysis. msPLSDA utilizes multiset sparse Partial Least Squares Discriminant Analysis to simultaneously extract disease-relevant features from multiple data modalities and predict disease states within a unified and supervised framework.
To demonstrate the effectiveness of msPLSDA, we conducted a comprehensive simulation study comparing it with existing methods. The results clearly indicate that msPLSDA outperforms other approaches in feature selection and discriminant analysis.
Furthermore, we applied msPLSDA to multi-omics data from the Louisiana Osteoporosis Study. Through this analysis, we identified genomic features that are significantly enriched in bone-related biological pathways. These findings underscore the effectiveness of msPLSDA in identifying critical features for downstream analyses, including the identification of disease biomarkers or therapeutic targets.
Contact: kmsh410 " at" hotmail "dot" com.
To get started with msPLSDA, please refer to the DataSimulationStudy_msPLSDA.R for the usage of the msPLSDA.
2018_msPLS-master: Directory containing the original code and files from the 2018 msPLS project.DataSimulationStudy_msPLSDA.R: R script for the data simulation study for msPLSDA.LICENSE: License information for the msPLSDA repository.README.md: This file, providing an introduction and instructions for using msPLSDA.RealDataAnalysis: Directory for storing files related to reading and analyzing data using msPLSDA.figure: Directory to store figures and images.
msPLSDA is released under the MIT License, granting users the freedom to use, modify, and distribute the software. However, please note that this repository includes third-party dependencies, each of which may have its own licensing terms. Please review the licenses of the respective dependencies before using this software.
We hope that msPLSDA will be a valuable tool for researchers and practitioners in the field of multi-omics analysis. If you have any questions or need further assistance, please don't hesitate to reach out. Enjoy exploring the potential of msPLSDA for your own multi-omics investigations!