License
Copyright 2020 Riccardo CAPPUZZO
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
EmbDI
EmbDI is a Python library developed to perform Entity Resolution (ER) and Schema Matching (SM) tasks by employing word embeddings. To test how well does the model encode information, we also implemented a set of Embeddings Quality (EQ) tests.
The repository includes a main script and a collection of additional scripts used to generate the files needed to
perform the tasks specified by the user.
main.pyis used to train the embeddings and/or perform postprocessing (ER, SM, EQ) on them. Execution may be done in batch by reading all files in a directory or by passing a single configuration file on the command line. Configuration files are explained in a separate section. The input file must be an edgelist that uses a specific format described in its section.generate_tests.pyproduces the files needed to test embedding quality ("match attributes"MA, "match rows"MRand "match columns"MC).edgelist.pyis used to generate the lists of edges used to build the graph starting from a relational dataset. This script is used for backward compatibility. It can also export the graph in networkx format, so that it can be used in other applications.
Contacts
Contact riccardo [dot] cappuzzo [at] inria [dot] fr for any inquiries.
Directory tree
Due to the different files required to run the code, both input and output files are saved in a directory
labeled pipeline. All data input/output will assume the presence of a directory with this name.
The expected directory tree is the following:
root
├─EmbDI
|
├─pipeline
├── config_files
├── datasets
├── dump
├── edgelists
├── embeddings
├── experiments
├── generated-matches
├── info
├── matches
├── replacements
├── sim_files
├── test_dir
└── walks
Additional directories are present in the project directory: they contain additional (not required) code or are needed for preprocessing or analysis.
Resources and dataset used in the paper
The datasets used for our experiments were converted into edgelists after performing some preprocessing steps.
We uploaded additional resources to reproduce our results in a separated cloud folder at:
https://zenodo.org/records/7930461
The additional resources include:
- (partially preprocessed) base datasets.
- Their Entity Resolution and Schema Matching versions.
- Edgelists for both ER and SM versions.
- Ground truth files for ER and SM tasks.
- Test directories for the EQ task.
- Copies of the configuration files provided in this repository.
Configuration files, info files and ER match files were left in this repository in pipeline/config_files/default,
pipeline/info and pipeline/matches/default.
main.py
main.py allows to execute the full pipeline (graph and corpus generation, embeddings training and matching), or a
subset of the pipeline steps depending on the parameters found in the task field present in the configuration file.
The script requires one of two command line parameters:
-d,--config-dirrequire the path to a directory that should contain configuration files. All valid files will be executed in alphabetical. All files present in the provided directory that do not start with the stringdefaultand that are not directories themselves will be considered as "valid".
python main.py -d path/to/config/dir/
-f,--config-fileshould be used to run only a single configuration file.
python main.py -f path/to/config/file
Configuration files are described in their own section. The task field is mandatory, as it specifies what operations
should be performed.
The possible tasks are the following:
train: train new embeddings using the parameters supplied in the configuration file and save them on the given output file. No further operations are performed on the output file.test: given a complete embeddings file (in text form), perform the tests (either EQ, ER, SM) specified in the configuration file.match: given a complete embeddings file (in text form), execute ER and SM and save on a file the matches, without testing their correctness. These matches can then be used for other tasks.train-test: train new embeddings and immediately run tests on them.train-match: train new embeddings and immediately produce matches.
Configuration files
Some example configuration files are stored in the folder pipeline/config_files/demo. An example of generic
configuration file is reported below.
Please note that the order of lines is not important, but the first field (before the :) has to be written how it is
shown here (unknown strings will raise exceptions).
All lines starting with # will be treated as comments and will be ignored. Similarly, all text following a # will be
ignored.
In the example below, lines starting with * are optional and, if missing, will be assigned a default value.
Values enclosed in {} denote parameters that require only one choice out of the proposed values.
# Input configuration:
task: {train, test, match, train-test, train-match}
input_file: path/to/edgelist/edgelist.txt
output_file: name_of_output_file # the final embeddings file will be saved in pipeline/embeddings/name_of_output_file.emb
experiment_type: {EQ, ER, SM}
match_file: path/to/match/file.txt [required for SM, ER]
dataset_file:path/to/dataset/file.csv [required for SM]
test_dir: path/to/EQ/tests/ [required for EQ]
embeddings_file: path/to/embeddings/file.emb [This field will be ignored if the task requires training]
walks_file: path/to/walks/file/file.walks [A valid walks file will bypass the graph and walks generation]
# Walks configuration:
*sentence_length: length_of_sentences
*n_sentences: {(int) number_of_sentences, default} # default will compute a "good" number of sentences based on the number of nodes in the graph
*follow_sub: {true, false} [If the strategy is Replacement, when a substitute is found, jump on the substitute.]
*smoothing_method: {no, smooth, inverse_smooth, log, piecewise} [Refer to the section on smoothing for more details]
*backtrack: {True, False} [Allow backtracking, i.e. given step n in a RW, it is possible to go back to the node in step n-1]
*write_walks: {True, False} [If True, walks will be dumped on drive. This will take marginally longer and reduce the memory cost.]
*repl_strings: {True, False} [If True, a similarity file will be needed to perfrom replacement]
*repl_numbers: {True, False}
*flatten: {(str) prefix_of_nodes_to_flatten, all, ''} [The graph algorithm will split all nodes with a prefix listed here]
# Embeddings configuration:
*learning_method: {CBOW, skipgram}
*window_size: w_size # Size of the context window in the word2vec training step
*n_dimensions: n_dim # Number of dimensions to be used for the final embeddings
# Test configuration:
*ntop: n_closest_neighbors_to_study
*ncand: n_candidates_from_ntop to take when looking for the closest. Boosts recall, reduces precision a lot
*max_rank: cutoff_rank_for_SM
# Miscellaneous:
*indexing: {basic, ngt, faiss, annoy} # Choose which indexing method to use in the ER task. Normally unnecessary, unless the embeddings are *very* large.
*epsilon: [parameter used by the ngt indexer]
*num_trees: [parameter used by the annoy indexer]
Default values
Default values are hardcoded in EmbDI.utils.py. Their values are the following:
'ntop' = 10
'ncand' = 1
'max_rank' = 3
'walks_file' = ''
'follow_sub' = False
'smoothing_method' = 'no'
'backtrack' = True
'training_algorithm' = 'word2vec'
'write_walks' = True
'flatten' = ''
'indexing' = 'basic'
'epsilon' = 0.1
'num_trees' = 250
'compression' = False
'n_sentences' = 'default'
'learning_method' = 'skipgram'
'window_size' = 3
'n_dimensions' = 300
Smoothing parameters
Experiments have shown that weighted random walks may perform better than regular random walks. Multiple weighing
functions are proposed and can be accessed by changing the smoothing_parameter in the configuration file.
The smoothing_parameter field should be a string that contains the smoothing method and (if needed) its parameters.
If only the name of the smoothing method is provided, default parameters will be used. The optional parameters will
be shown in [].
Implemented smoothing methods
no: default strategy, all nodes will have the same weight and will therefore be chosen with the same likelihood.smooth,[k=0.2, target=200]: inverse exponential function. The slope of the exponential can be tweaked by changing the value oftarget(larger targed = lower slope). The exponential function will also approximate the valuek, rather than 0. This is done to prevent situations where some nodes will never be reached. This function will penalize frequent nodes, assigning them a weight ofk.inverse_smooth, [s=0.1]: a flipped exponential function that will approximate weight 1 as the frequency of a node increases. In this case, the minimum weight that can be assigned to a node is0.5for nodes with frequency 1. We observed that this function behaves better thansmoothwhen applied to some datasets. The slope of the function can be set with the parameters, a smaller value ofswill reduce the slope. In this case, we advise to test cases withs= 0.1, 0.01... as the function increases very quickly.
Example of tuple:
smoothing_method: smooth,0.5,200
This will use the smooth function with k=0.5 and target=200 .
Preparing the edgelist
EmbDI trains embeddings by building a graph on the basis of an edgelist supplied by the user.
The file edgelist.py generates an edgelist given a csv file. It takes two required arguments from the command
line:
-i INPUT_FILE, --input_file INPUT_FILE Path to input csv file to translate.-o OUTPUT_FILE, --output_file OUTPUT_FILE Path to output edgelist.
The format of the edgelist is one of the following:
n1,n2,w1,w2>> node1, node2, weight1to2, weight2to1n1,n2>> node1, node2, 1, 1n1,n2,w1>> node1, node2, weight1to2 [no link back, directed graph]
Edgelist prefixes
The first line of the edgelist must contain all the prefixes that define node types (such as RID, CID, tokens). These prefixes will be used to tell the algorithm how to handle the different node types. During our experimental campaign, we noticed that variations in how nodes are represented in the random walks were reflected in the performance of the algorithm in the different tasks. For this reason we implemented the following prefix/class system, as it would allow us to have a more systematic method for customizing these representations.
The default set of prefixes used in the edgelists generated for EmbDI is , where each
comma-separated value has the specific format [1-7][#-$]__node_type_name:
- The first character should be a numeric value in the range
1-7, which will denote theclassof the node type. - The second character is used to distinguish nodes that contain numeric values (with symbol
#) from those that contain categorical information ($).
The class will influence the behavior of the random walks generation algorithm.
There are 7 possible classes based on the truth table reported below. first means that the node may be chosen
as first value in a sentence. root means that the node will be added to the pool of nodes to start from when
generating sentences. appears means that, when the random walks hits the node, the node will appear in the
sentence, otherwise it will not be saved.
Example of prefix with explanation
Considering the example 3#__tn,3$__tt,5$__idx,1$__cid:
- Nodes with prefix
tn__will be treated as numbers (because of the#character) and undergo some additional processing in the graph generation phase. - Nodes with prefix
tt__,idx__andcid__will be treated as strings (due to character$). tt__andtn__nodes are in class 3. Nodes in class 3 will be used as root for the random walks (that is, paths will begin from nodes in this class). This means that the random walks generation algorithm will build the training corpus by going through all nodes marked as "root" and generate a certain number of random walks starting from each of them.idx__nodes are in class 5. In our experiments, we noticed that Entity Resolution results improved when random walks featured an additionalidx__node before the root. Given a random walktt__G2, idx_0, tt_A3, cid__col1, its "improved" version according to this method (denoted asfirst) would beidx_3, tt_G2, idx_0, tt_A3, cid__col1cid__nodes are in class 1. In some training scenarios, the presence of a node clas in the training corpus would make training more difficult and produce worse representations of the nodes, which in turn caused issues down the line. Tasks that revolved entirely around cell value embeddings would be less successful because of the presence of additional nodes that "diluted" the amount of information available in the training corpus. To mitigate this issue, it is possible to mark some nodes using theappearflag to signal to the algorithm that a certain class of nodes should be considered as a viable neighbor when generating each random walk, but at the same time nodes belonging to that class should not appear in the training corpus. For example, in a case in which bothidx__andcid__nodes have "appear" set to false, the random walktt__G2, idx_0, tt_A3, cid__col1, tt_G5, idx_0 ...would be saved on disk as:tt__G2, tt_A3, tt_G5, ...
| Class | First | Root | Appears |
|---|---|---|---|
| 0 | - | - | - |
| 1 | - | - | + |
| 2 | - | + | - |
| 3 | - | + | + |
| 4 | + | - | - |
| 5 | + | - | + |
| 6 | + | + | - |
| 7 | + | + | + |
It is possible to flatten all nodes using flatten:all in the configuration file. Alternatively,
nodes can be flattened based on their type by listing the prefixes to expand separated by a comma.
For example, if the prefixes are tt, tn, idx, cid like in the standard csv-translation case, by putting flatten:tt only the
nodes with prefix tt will be flattened.
Additional files
Matches file
The matches file is the ground truth to be used when performing ER or SM tasks. The system will expect a known format for the ground truth.
Below is shown an example for the ER case. Each line should contain a match between a tuple in one dataset and one tuple in the other dataset written as "idx__[line_id]". The line_id value refers to the line number of the tuple in the dataset passed as input to the training algorithm, after the concatenation of the original datasets.
Note: the indices start from idx__0 (rather than idx__1), so in a typical spreadsheet application idx_0 will correspond to line 2 because of the header and the fact that row indices start from 1, rather than from 0.
Multiple matches are allowed, but the current code implementation will only find the closest one.
...
idx__1007,idx__13670
idx__1007,idx__20696
idx__1009,idx__3334
idx__1010,idx__17557
idx__1011,idx__15967
idx__101,idx__5147
idx__1014,idx__9582
idx__102,idx__14313
...
The matches file for the SM case (here, for the DBLP_Scholar dataset) looks like this:
0_authors,1_authors
0_title,1_title
0_venue,1_venue
0_year,1_year
SM tasks take as inputs datasets whose columns were renamed to "label_dataset_n_[column_name]". For example,
the header of the DBLP-ACM dataset will become
0_author_1,0_author_2,0_author_3,0_author_4,0_title,0_venue,0_year,1_author_1,1_author_2,1_author_3,1_author_4,1_title,1_venue,1_year.
To properly concatenate the dataset, it is possible to use the data_preprocessing model provided here using the horizon
parameter.
All following lines should contain the ground truth matches. If a column can be matched arbitrarily to multiple columns, multiple matches are allowed. As in the ER case, however, only the closest ones will be considered. This behavior will change in a future version of the code.
Note on results
It may happen that results differ slightly between different runs. In our experience, there may be some variance in the ER results because of the random walks generation, as well as the embeddings training procedure.
In SM, the difference might become larger due to the fact that there are only a few "matches", so even a single miss will drop the precision by a large amount.
We observed that different random walks lead to slightly different embeddings. In our matching algorithms we use k-nn to find the best candidates, and the ranking may be influenced by how the embeddings are positioned in different runs.
Similarity file
Currently under work, unavailable. The similarity file contains candidates for replacement with the structure, optionally with their similarity in the range [0,1]. If no distance is supplied, it is assumed to be 1 (word1 == word2).
...
word1_dataset1,word1_dataset2,similarity1
word2_dataset1,word2_dataset2,similarity2
...
Example:
...
english,en, 0.8
french,fr, 0.8
italian,it, 0.8
...
Generation of tests
EQ tests can be generated by using the generate_test.py script. In its current implementation, parameters are hardcoded
and must be changed in the script itself.
Three different tests are currently provided:
- nmr (no-match-rows, MR in the paper) tests observe whether the provided embeddings are able to recognize what value within a tuple was exchanged with a value that does not belong to that tuple.
- nmc (no-match-column, MA in the paper) tests observe whether the embeddings can find what value does not belong to a given attribute
- nmcon (no-match-concept, MC in the paper) tests observe if a semantic link between values is stronger than the link between values belonging to the same attribute (e.g. given director Steven Spielberg and target values Jurassic Park, Saving Private Ryan, Jaws and Star Wars: A new Hope, nmcon tests expect Star Wars as the answer, since it was not directed by Steven Spielberg).
How tests are generated
For NMR tests, n_sentences tests will be generated for all attributes in the list supplied to nmr_attrs_to_change. For
all such values, n_sentences random tuples will be chosen (with replacement) and the value corresponding to the
target attribute is substituted by a different value. The test is successful if the new value is selected.
In NMC tests, n_sentences tests will be prepared for all combinations of attributes found in the list
nmc_col_combinations. For each pair of attributes, a random selection of unique values coming from the first attribute is
extracted, and then a random value from the second attribute is selected. The test is passed if the testing algorithm
chooses the value from the second attribute as its answer.
In NMCON tests, pairs supplied in the list fd will be tested as by finding those values in the first attribute that
have more than test_length distinct values in the second column (e.g. given pair director, movie_title and
test_length=5, only those directors found in 5 or more tuples with distinct movie titles will be chosen). Then,
tests are generated by picking a random eligible value, test_length-1 related values and one unrelated value. Finally,
the test is passed if the unrelated value is chosen by the EQ algorithm.
Data Preprocessing
The Data Preprocessing module can be used to process the datasets to be tested. It includes the following features:
- Data exploration by column
- Removal of missing values (the user can provide additional examples)
- Multiple strategies to handle missing values
- Automatic rounding of columns
- Automatic merging of similar words
check_info(df, columns)
A simple exploration per column:
- number of not null values
- number of null values
- number of unique values
- number of duplicated values
- number of duplicated instances
data_preprocessing(dfs, params)
The main data process function. It takes as input a list of datasets to preprocess and returns the concatenated dataset with the application of all preprocessing steps. The supported preprocessings steps are the following:
- normalization of missing values: the given strings will be treated as missing values.
- normalization of text values: strings are set to lowercase, trailing spaces are removed, some special characters are removed, words are concatenated into one single string.
- rounding of numerical fields: numerical fields (user-supplied, if needed) are rounded to the given number of digits after the decimal point
- concatenation of datasets: the input datasets are concatenated using one of the given methods:
{horizon,inner,outer}horizonwill concatenate the datasets in such a way that no column in the first dataset will contain values coming from the second dataset (they will be replaced by NULLs). This is the format required to perform Schema Matching.innerwill concatenate common columns and drop columns that are not present in both datasets.outerwill concatenate common columns and keep the remaining columns, filling missing values with NULLs.
- token merging: similar tokens (according to multiple similarity measures) are merged into one to increase the amount of overlap between the two tables. MinHash LSH merging is supported. Tokens that satisfy the LSH threshold and the distance threshold are merged into a new one.
- replacement of missing values: three strategies are provided to replace missing values
{'separated_null', 'one_null', 'ignore'}.
Default params:
parameters = {
'missing_value': 'nan,ukn,none,unknown,',
'missing_value_strategy': '',
'round_number': -1,
'round_columns': '',
'concatenate': '',
'auto_merge': False,
'mh_k_shingles': 3,
'mh_threshold': .5,
'mh_perm': 128,
'distance': 'normalized_edit_distance',
'distance_threshold': .20,
'merge_columns': ''
}
Example of use:
parameters = {
'round_number': 0,
'round_columns': 'price',
'auto_merge': False
}
df_c = data_preprocessing([df1, df2], parameters)
LSHMerge
A class used to handle token merging. It is used as a first blocking step to find merge candidates.
Multiple user-defined parameters are supported.
The merge process is done in two steps:
LSH MinHash: Neighbors are found and placed in buckets. Params:k-shingles: group characters by k-shingles or by word. Shingles separate words in substrings and index them independently.minhash threshold: the similarity threshold to decide whether two items are similar or not.perm: number of MinHash permutations.
Distance metric: Once the buckets are ready, the distance metric is applied to all points inside the same bucket. For the moment, two distance metrics are provided.edit_distance: the distance will measure the number of characters to edit, d >= 0normalized_distance:edit_distance/len(str), d in [0, 1]none: do not use a distance function, rely only on LSH buckets.
We suggest not to rely on the default values and to observe the replacement candidates before replacing them: the edit distance does not take into consideration semantic closeness, so it may happen that unrelated terms will be merged in the same token. A conservative distance value should be employed.
Default params:
k_shingles=3
mh_threshold=.5
mh_num_perm=128
delimiter='_'
Example of use:
uniq_values = set(['white', 'whitte', 'wwhite'])
lsh = LSHMerge(uniq_values, 4, .5, 128)
# find neighbors of 'white'
neighbors = lsh.get_similarities('white')
# list out 5 random bins
lsh.get_sample_blocks(5)
# get the final replacement list to do the merge
replacement = lsh.get_replacement('normalized_edit_distance', .4)
Preparing datasets for the SM task
To perform Schema Matching, EmbDI will expect datasets formatted according to a specific structure.
Columns should be renamed to unique values (e.g. [0_author_1, 0_author_2] and [1_author_1, 1_author_2]) in such a
way that the concatenation does not put together values from different datasets. This can be achieved by setting the
parameter concatenate: horizon in the data preprocessing pipeline explained above.
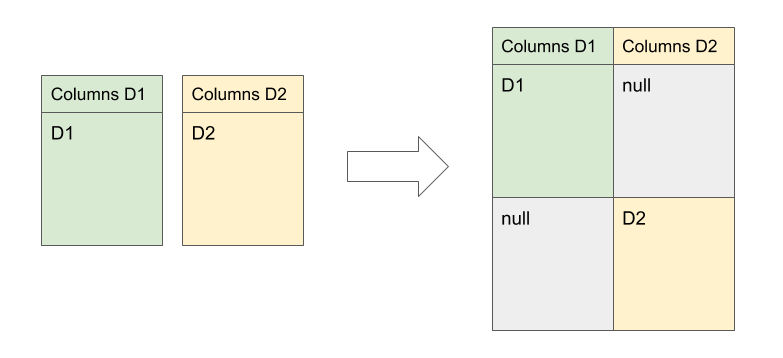
The result will be a dataset in which all tuples belonging to Dataset 1 will contain values in Dataset 1's columns, and nulls in Dataset 2's columns; similarly, tuples in D2 will contain nulls in D1's columns, and the proper values in D2's columns.
Reference
If you use EmbDI in your work, please cite it as follows:
@article{Cappuzzo_2020,
title={Creating Embeddings of Heterogeneous Relational Datasets for Data Integration Tasks},
ISBN={9781450367356},
url={http://dx.doi.org/10.1145/3318464.3389742},
DOI={10.1145/3318464.3389742},
journal={Proceedings of the 2020 ACM SIGMOD International Conference on Management of Data},
publisher={ACM},
author={Cappuzzo, Riccardo and Papotti, Paolo and Thirumuruganathan, Saravanan},
year={2020},
month={May}
}