Note: The Fibrosis-Net model provided here is intended as a reference model that can be built upon and enhanced as new data becomes available. It is currently at a research stage and not a production-ready model (not meant for direct clinical usage). We are working continuously to improve it as new data becomes available. Please do not use Fibrosis-Net for self-diagnosis and seek help from your local health authorities.
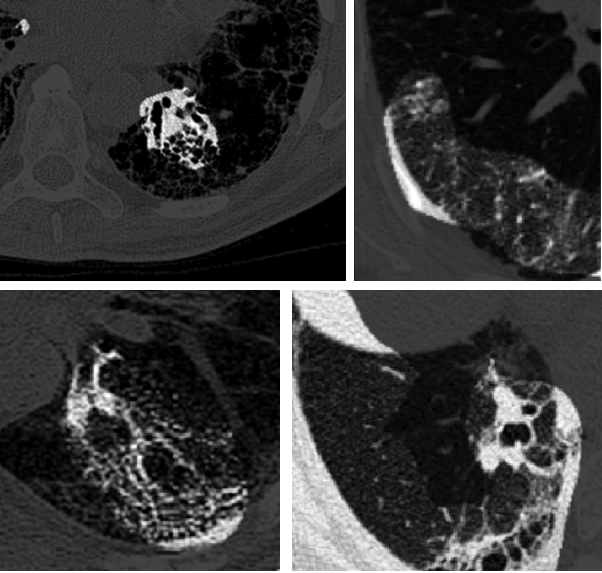
Example images and their associated critical factors (highlighted in white) as identified by GSInquire.
Pulmonary fibrosis is a devastating chronic lung disease that causes irreparable lung tissue scarring and damage, resulting in progressive loss in lung capacity; it has no known cure. A critical step in the treatment and management of pulmonary fibrosis is the assessment of lung function decline. Computed tomography (CT) imaging is a particularly effective method for determining the extent of lung damage caused by pulmonary fibrosis. Motivated by this, we introduce Fibrosis-Net, a deep convolutional neural network design tailored for the prediction of pulmonary fibrosis progression from chest CT images. More specifically, we leveraged machine-driven design exploration to determine a strong architectural design for CT lung analysis, upon which we built a customized network design tailored for predicting forced vital capacity (FVC) based on a patient's CT scan, initial spirometry measurement, and clinical metadata. Finally, we leveraged an explainability-driven performance validation strategy to study the decision-making behaviour of Fibrosis-Net and verify that predictions are based on relevant visual indicators in CT images. Experiments using the OSIC Pulmonary Fibrosis Progression Challenge benchmark dataset showed that the proposed Fibrosis-Net is able to achieve a significantly higher modified Laplace Log Likelihood score than the winning solutions on the challenge leaderboard. Furthermore, explainability-driven performance validation demonstrated that the proposed Fibrosis-Net exhibits correct decision-making behaviour by leveraging clinically-relevant visual indicators in CT images when making predictions on pulmonary fibrosis progress. Fibrosis-Net is available to the general public in an open-source and open access manner as part of the OpenMedAI initiative. While Fibrosis-Net is not yet a production-ready clinical assessment solution, we hope that releasing the model will encourage researchers, clinicians, and citizen data scientists alike to leverage and build upon them.
For a detailed description of the methodology behind Fibrosis-Net and more information, please click here.
If you find our work useful, you can cite our paper using:
@misc{wong2021fibrosisnet,
title={Fibrosis-Net: A Tailored Deep Convolutional Neural Network Design for Prediction of Pulmonary Fibrosis Progression from Chest CT Images},
author={Alexander Wong and Jack Lu and Adam Dorfman and Paul McInnis and Mahmoud Famouri and Daniel Manary and James Ren Hou Lee and Michael Lynch},
year={2021},
eprint={2103.04008},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
- Fibrosis-Net is comprised of two main components,
FibrosisNet-CTandFibrosisNet-Clinical. TheFibrosisNet-CTcomponent takes in the provided CT scans along with clinical metadata, while theFibrosisNet-Clinicalcomponent takes in the provided clinical metadata. - A pre-trained model,
FibrosisNetCT.pb, is included in the "models/" directory.
To reproduce our results for Fibrosis-Net, first clone this repository, and download the dataset here.
After extraction of the code and dataset, navigate into the main folder and install dependencies using:
pip3 install -r requirements.txt
Now you can run the following code to generate our Kaggle submission file. Make sure to modify the arguments as necessary. Running the code using CT_WEIGHT=1.0 achieves -6.8188 private score. Running it with CT_WEIGHT=0.96 achieves -6.8195 private score. Note that the DarwinAI team have reported a 0.0001 deviation in score between different Kaggle accounts and days of testing.
python3 kaggle_submission.py --datapath="osic-pulmonary-fibrosis-progression/" --modelpath="models/" --ctweight=1.0 --outputfile="submission.csv"
The main requirements are listed below. A full list can be found in "requirements.txt".
- Tested with Tensorflow 1.15
- OpenCV 4.5.1
- Python 3.6
- Numpy 1.20.0
- Pandas 1.2.1
In order to perform custom evaluation using the evaluation script, you must first run the "kaggle_submission.py" script (this automatically generates the additional models required for evaluation).
Set up the evaluation inputs. Refer to the directory "example_input" for an example. There must be a .csv file inside, with these columns:
- Patient
- Weeks
- FVC
- Percent
- Age
- Sex
- SmokingStatus
- PredictWeek
For each patient listed in the .csv file, a folder containing CT scan images should be available in the same directory. Each folder should correspond to a patient in the .csv file, linked by folder names.
Run the following code and make sure to modify the arguments accordingly. If successful, a .csv file will be generated, containing all of the predictions for 100 weeks for each patient.
python3 fibrosisnet_eval.py --modelpath="models/" --inputpath="example_input/" --ctweight=1.0 --outputfile="results.csv"
