Ozymandias
Ozymandias is a biodiversity knowledge graph, and was originally created as an entry into the 2018 GBIF Ebbe Nielsen Challenge. For many people biodiversity data is a taxonomic name attached to a specimen or observation that can be placed on a map. Typically this data is stored in tables and viewed as lists or maps. Existing biodiversity databases rarely link to the scientific research underlying their contents, such as the taxonomic literature. Hence the data is disconnected from supporting evidence, and from the researchers who gathered that evidence.
There is a growing volume of open data coming from sources such as taxonomic databases, digital libraries, genomics, and wiki projects. To make the best use of this data we need to move beyond tables to thinking in terms of connected networks of relationships, i.e. knowledge graphs. Knowledge graphs link different kinds of data together using shared identifiers, such as DOIs (e.g., for articles), LSIDs (e.g., for taxonomic names), ORCIDs (for people), and UUIDs (for anything). By linking data and displaying the data and its connections we can create rich experiences for casual users, students, and researchers alike. Any entity in the knowledge graph can be the focus of investigation. You can focus on what we know about a particular species, or explore the activities of a researcher, or discover the output (journals, articles, taxonomic descriptions) associated with a particular institution. Hence we could also use a knowledge graph to inform data collection and management policies, for example, by discovering gaps in literature digitisation, or uneven representation of content from different institutions. WE could help boostrap the engagement of researchers in data curation by avoiding having to ask them to demonstrate their expertise - if a researcher has an ORCID we can discover their list of publications and the taxa they work on.
Ozymandias is a live example of a knowledge graph https://ozymandias-demo.herokuapp.com. Given the constraints of the challenge, this knowledge graph is limited to taxa, publications, researchers, journals, and instutitions, and the taxonomic scope is the animals in the Atlas of Living Australia. In future I hope to add other eukaryote taxa, and extend the graph to include specimens and sequences.
Model
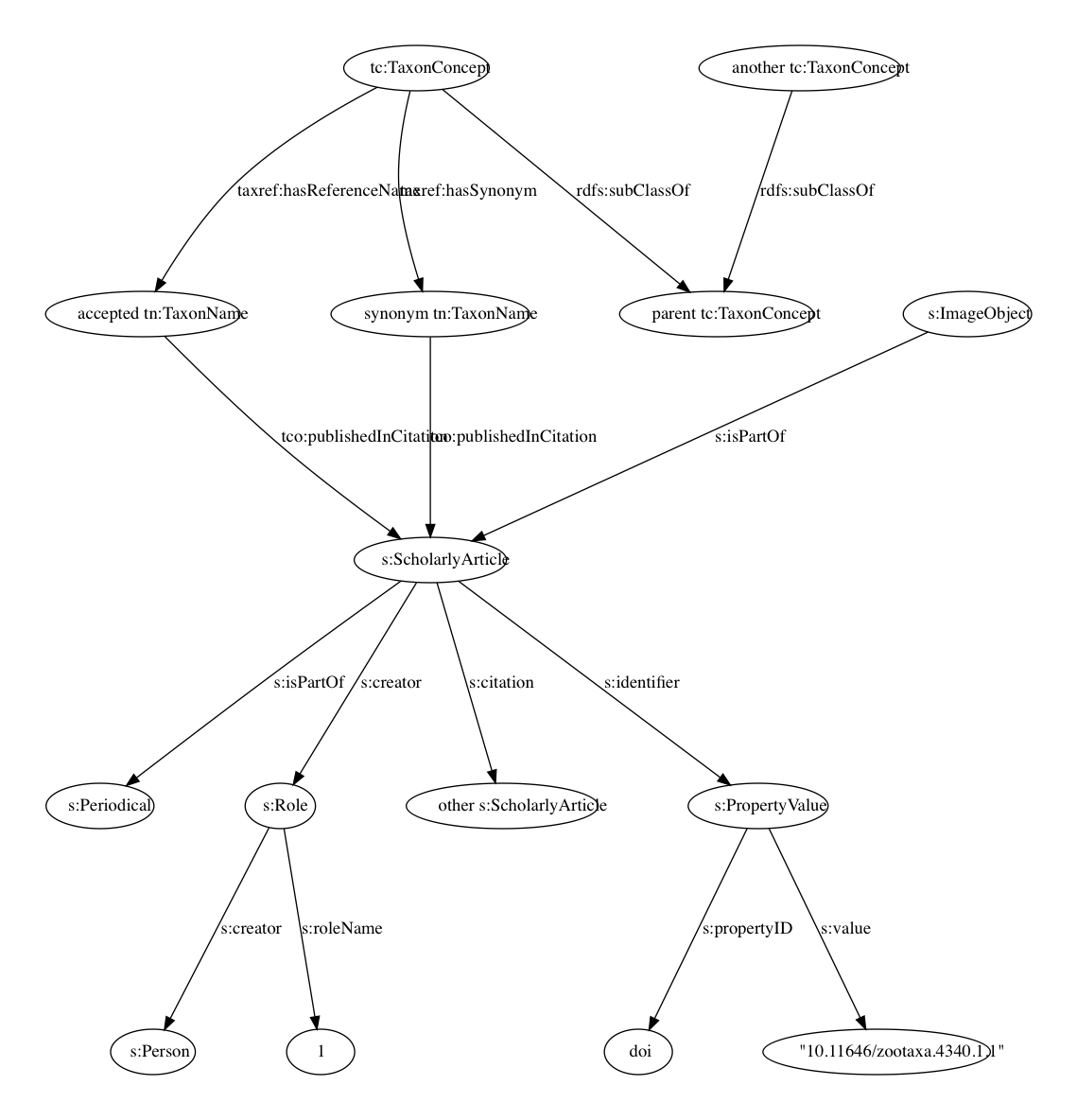
Below is a simplified model of the knowledge graph. The core entities are taxa, taxonomic names, publications, journals, and people.
Taxa have type http://rs.tdwg.org/ontology/voc/TaxonConcept#TaxonConcept. A taxonomic classification is represented by rdfs:subClassOf relationship between parent and child taxa (a child is a rdfs:subClassOf its parent). Taxa are connected to taxonomic names (type http://rs.tdwg.org/ontology/voc/TaxonName#TaxonName) by relations from the taxref vocabulary, and are typically either accepted names or synonyms. Names are published in publications (typically of type schema:ScholarlyArticle but may also be other types derived from schema:CreativeWork). Articles are schema:isPartOf journals (schema:Periodical). Authors are linked to their publications using “roles” (schema:Role) which enables us to include information on order of authorship. Publications may be linked by schema:citation relations. Figures within a publication (schema:ImageObject) are schema:isPartOf that publication. To handle the existence of multiple identifiers I create a schema:PropertyValue item for each identifier (linked to the publication using schema:identifier) and store the identifier (stripped of any resolution mechanism) as a schema:value. This indirection avoids having to figure out which IRI is used to identify an entity, instead of asking for the entity with IRI https://doi.org/10.11646/zootaxa.4340.1.1 (or some variation of that URL) we can ask what entity as an identifier with schema:propertyID “doi:” and schema:value “10.11646/zootaxa.4340.1.1”.
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?root_name ?parent_name ?child_name WHERE
{
VALUES ?root_name {"HYDROPTILIDAE"}
?root <http://schema.org/name> ?root_name .
?child rdfs:subClassOf+ ?root .
?child rdfs:subClassOf ?parent .
?child <http://schema.org/name> ?child_name .
?parent <http://schema.org/name> ?parent_name .
}
http://130.209.46.63/blazegraph/sparql
Server notes
Windows 10
Hosting on local Windows 10 PC (to avoid cost of cloud hosting).
Blazegraph
Need Java version 7, which can be obtained from Oracle.
To start:
java -server -Xmx4g -jar blazegraph.jar
This runs on port 9999 so we use nginx as a reverse proxy (see below).
If loading times are getting very slow, specially when reloading data and experimenting you may want to start from scratch. To do this stop the server, delete the file blazegraph.jnl and restart blazegraph.
nginx
I use nginx to act as reverse proxy for Blazegraph running on Windows.
# forward to Blazegraph listening on 127.0.0.1:9999
#
location /blazegraph {
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header Host $http_host;
proxy_pass http://127.0.0.1:9999;
}
When uploading data I often got HTTP 413 Request Entity Too Large errors, which can be fixed by setting client_max_body_size to a suitable value in the server part of nginx.conf file, for example:
client_max_body_size. 200M;
Another problem is the server timing out if Blazegraph is doing a task which takes a while (HTTP 504). To fix this and these settings to the http section:
proxy_connect_timeout 600;
proxy_send_timeout 600;
proxy_read_timeout 600;
send_timeout 600;
(See How to Fix 504 Gateway Timeout using Nginx).
Firewall
Need to add nginx to the Windows Firewall rules so that it can be accessed by the outside world.
Sloppy.io
To host knowledge graph on Sloppy.io use openkbs/blazegraph with 8GB of RAM and 3 volumes (24 GB in total):
{
"project": "kg",
"services": [
{
"id": "blazegraph",
"apps": [
{
"id": "openkbs",
"image": "openkbs/blazegraph",
"instances": 1,
"mem": 8192,
"domain": {
"uri": "kg-blazegraph.sloppy.zone"
},
"ssl": false,
"port_mappings": [
{
"container_port": 9999
}
],
"volumes": [
{
"container_path": "/data",
"size": "8GB"
},
{
"container_path": "/home/developer/blazegraph/conf",
"size": "8GB"
},
{
"container_path": "/home/developer/data",
"size": "8GB"
}
],
"health_checks": [
],
"logging": null
}
]
}
]
}
Digital Ocean
8 Gb droplet
Create a droplet.
docker-machine create --digitalocean-size "s-4vcpu-8gb" --driver digitalocean --digitalocean-access-token xxx ozymandias
eval $(docker-machine env ozymandias)
#### Blazegraph
docker run -d -p 9999:9999 openkbs/blazegraph
## Other notes
Beyond classifying people as researcher/non-researcher https://twitter.com/SiobhanLeachman/status/1025203488102334464
## Dump data
Dump all the data from the triple store. Get the named graphs:
SELECT ?g WHERE { GRAPH ?g { } }
Then export each named graph and save in a separate file:
oz-ala.nt https://bie.ala.org.au oz-publication.nt https://biodiversity.org.au/afd/publication oz-zenodo.nt https://zenodo.org oz-crossref.nt https://crossref.org oz-orcid.nt https://orcid.org oz-species.nt https://species.wikimedia.org oz-gbif.nt https://gbif.org/species oz-bold.nt http://boldsystems.org
```curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <https://bie.ala.org.au> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-ala.nt
curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <https://biodiversity.org.au/afd/publication> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-publication.nt
curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <https://zenodo.org> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-zenodo.nt
curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <https://crossref.org> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-crossref.nt
curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <https://orcid.org> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-orcid.nt
curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <https://species.wikimedia.org> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-species.nt
curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <https://gbif.org/species> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-gbif.nt
curl -X POST http://130.209.46.63/blazegraph/sparql --data-urlencode "query=CONSTRUCT { ?s ?p ?o } WHERE { graph <http://boldsystems.org> { hint:Query hint:constructDistinctSPO false . ?s ?p ?o } }" -H 'Accept:text/x-nquads' > oz-bold.nt
Examples, errors, etc.
Examples
Lots of papers
Wallacellus is Euwallacea: molecular phylogenetics settles generic relationships (Coleoptera: Curculionidae: Scolytinae: Xyleborini)
Three new species of Fergusonina Malloch gall-flies (Diptera: Fergusoninidae) from terminal leaf bud galls on Eucalyptus (Myrtaceae) in south-eastern Australia
Occurrences
Experimenting with adding GBIF occurrences, e.g. https://ozymandias-demo.herokuapp.com/?uri=https://gbif.org/occurrence/1100252191
https://ozymandias-demo.herokuapp.com/?uri=https://gbif.org/occurrence/1101089151
USNMENT809090 https://ozymandias-demo.herokuapp.com/?uri=https://gbif.org/occurrence/1317230794
BOLD ANICH163-10 https://ozymandias-demo.herokuapp.com/?uri=http%3A%2F%2Fboldsystems.org%2Findex.php%2FPublic_RecordView%3Fprocessid%3DANICH163-10%23occurrence
https://www.ncbi.nlm.nih.gov/nuccore/HQ245367.1 https://www.ncbi.nlm.nih.gov/nuccore/GU302250.1 https://ozymandias-demo.herokuapp.com/?uri=https://bie.ala.org.au/species/urn:lsid:biodiversity.org.au:afd.taxon:e69a24f8-a906-4bff-8776-c836f87aa4ad
Nice figures
https://ozymandias-demo.herokuapp.com/?uri=https://doi.org/10.5281/zenodo.189913
Multiple author names
Variation in author names causes problems, e.g. https://ozymandias-demo.herokuapp.com?uri=https://biodiversity.org.au/afd/publication/a7cc7f8d-7e09-4cc8-916c-423b21b19d98
- T. Y. Chan
- T.-Y. Chan
- T. Y Chan
- T-Y Chan
- T-Y. Chan
All due to missing “.” and “-“.