The goal of isoswitch is to facilitate the characterization of isoform
expression in long-read single-cell datasets. It includes a set of
functions and reports built on top of Seurat, ggplot and rmarkdown
that can be used to search, visualize and document isoform expression
patterns, and particularly isoform switches between cell identities.
You can install the development version of isoswitch from GitHub with:
# install.packages("devtools")
devtools::install_github("ucagenomix/isoswitch")- Input data setup & pre-processing
- Isoform characterization
- Isoform switch detection
- Gene reports
Below is a short overview of the package functionality:
Isoswitch is designed to work with Seurat objects with gene- and
isoform-level counts.
In particular, the ScNaUmi-seq protocol (Lebrigand et al 2020) generates
two
count matrices that need to be loaded into respective Seurat assays
before starting the analysis, more specifically:
- A gene-level [gene x cell] matrix count, generated by 10X CellRanger pipeline. Typically stored in assay “RNA” in Seurat object.
- An isoform-level [isoform x cell] matrix count generated by SiCeLoRe pipeline, stored in a separate “isoform” assay.
Note
isoswith expects the row names of the isoform count matrix to follow the format “[gene_name]..[transcript_id]”, example: “BCS1L..ENST00000359273”
head(rownames(seurat@assays$multi@counts))
#> [1] "BCS1L..ENST00000359273" "PPP1R10..ENST00000461593"
#> [3] "OSBPL9..ENST00000428468" "TEF..ENST00000406644"
#> [5] "CBX5..ENST00000209875" "TRAP1..ENST00000246957"After loading up gene and isoform counts on the Seurat object, the
method iso_preprocess() prepares the isoform matrix by removing
low-expression transcripts as well as removing single-isoform genes
which are irrelevant for the isoform switch analysis.
The resulting “multi-isoform” matrix is stored as a new assay on the input Seurat object.
seurat <- iso_preprocess(seurat, assay="ISO", new_assay="multi", filter_threshold=5)The method iso_compute_stats() parses the isoform raw count matrix
returning a data frame with stats on the expression of each transcript
feature,gene_id,transcript_idgene/transcript identifierssumTotal UMI counts for the transcripttotal_geneTotal UMI counts for the genen_isofsNumber of distinct isoforms detected for the genemax_sumMax sum value for the isoform with the highest expressionpercRelative percentage of isoform UMI count vs gene total.is_major(Boolean) is this considered a major isoformis_top(Boolean) is this highest expressed isoform
stats <- iso_compute_stats(seurat@assays$multi@counts) %>% arrange(gene_id)
head(stats, n=4)
#> feature gene_id transcript_id sum total_gene n_isofs max_sum
#> 1 A1BG..ENST00000596924 A1BG ENST00000596924 5 8 2 5
#> 2 A1BG..ENST00000598345 A1BG ENST00000598345 3 8 2 5
#> 3 A2M..ENST00000495709 A2M ENST00000495709 10 14 2 10
#> 4 A2M..ENST00000318602 A2M ENST00000318602 4 14 2 10
#> perc is_major is_top
#> 1 62.50000 TRUE TRUE
#> 2 37.50000 TRUE FALSE
#> 3 71.42857 TRUE TRUE
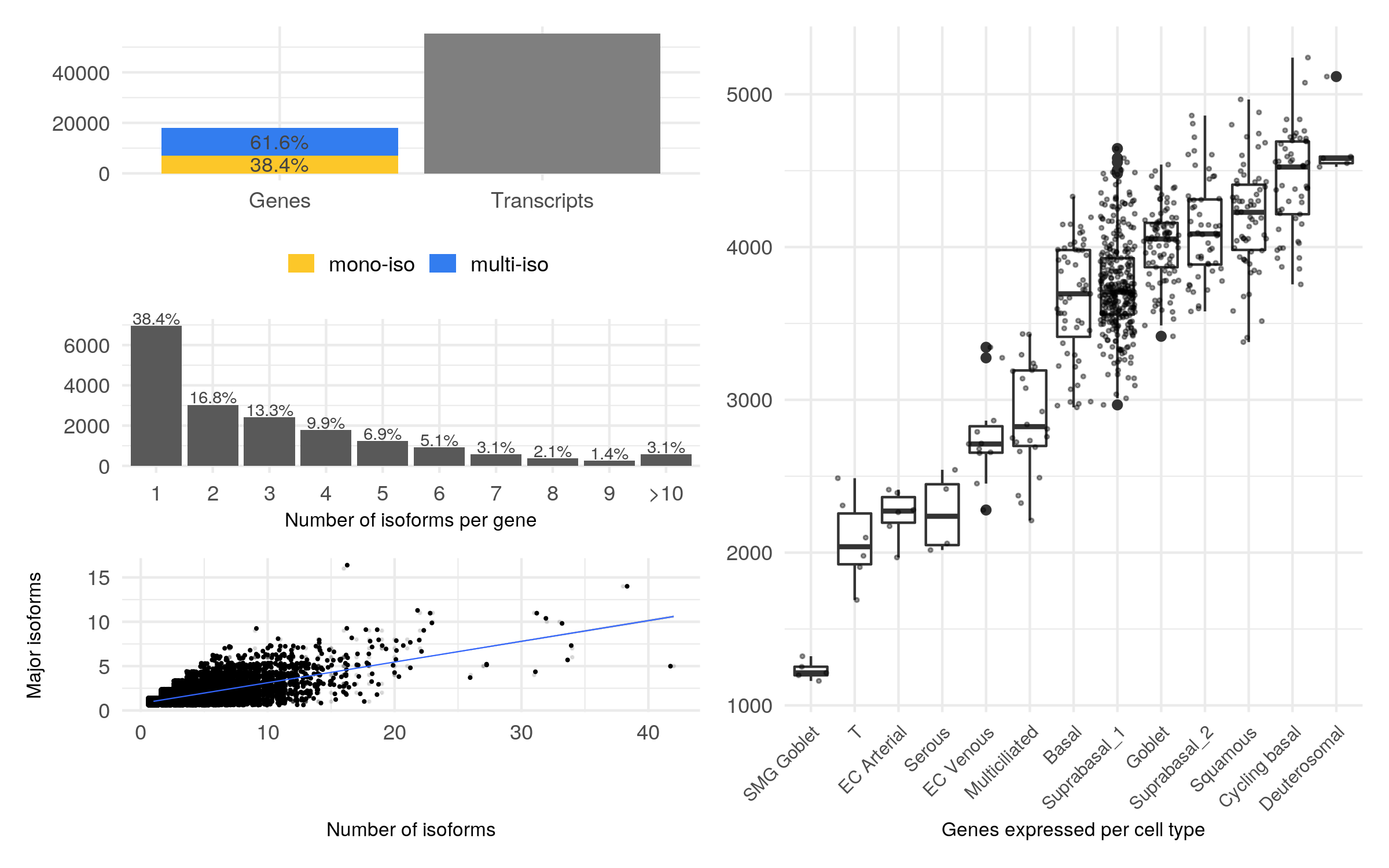
#> 4 28.57143 FALSE FALSEThe method plot_assay_stats() builds on this data to plot a summary
with number of genes, number of transcripts, distribution of isoforms
and number of genes per cell type that can be used to describe succintly
the isoform distribution in the dataset.
plot_assay_stats(seurat, "isoform")The term “isoform switch” refers to an event where two isoforms of the same gene are considered markers of different clusters.
The marker search is implemented on the method ISO_SWITCH_ALL(). Any
extra parameters are passed on to the underlying Seurat’s FindMarkers
call to fine tune the search space.
clusters <- levels(seurat@active.ident)
switch_markers <- ISO_SWITCH_ALL(seurat, clusters, assay="isoform",
min.pct=0, logfc.threshold=0.40)ISO_SWITCH_ALL() returns a data frame of transcripts identified as
markers of a given cluster for a given gene, one transcript per row.
To facilitate the graphical interpretation of the results:
plot_marker_matrix()produces a heatmap of number of unique genes per contrast between clustersplot_marker_score()produces a volcano plot showing p-values and average logFC for each gene with an isoform switch
pl1 <- plot_marker_matrix(seurat, switch_markers)
pl2 <- plot_marker_score(adult, switch_markers, facet=FALSE, overlaps=16)
pl1 | pl2 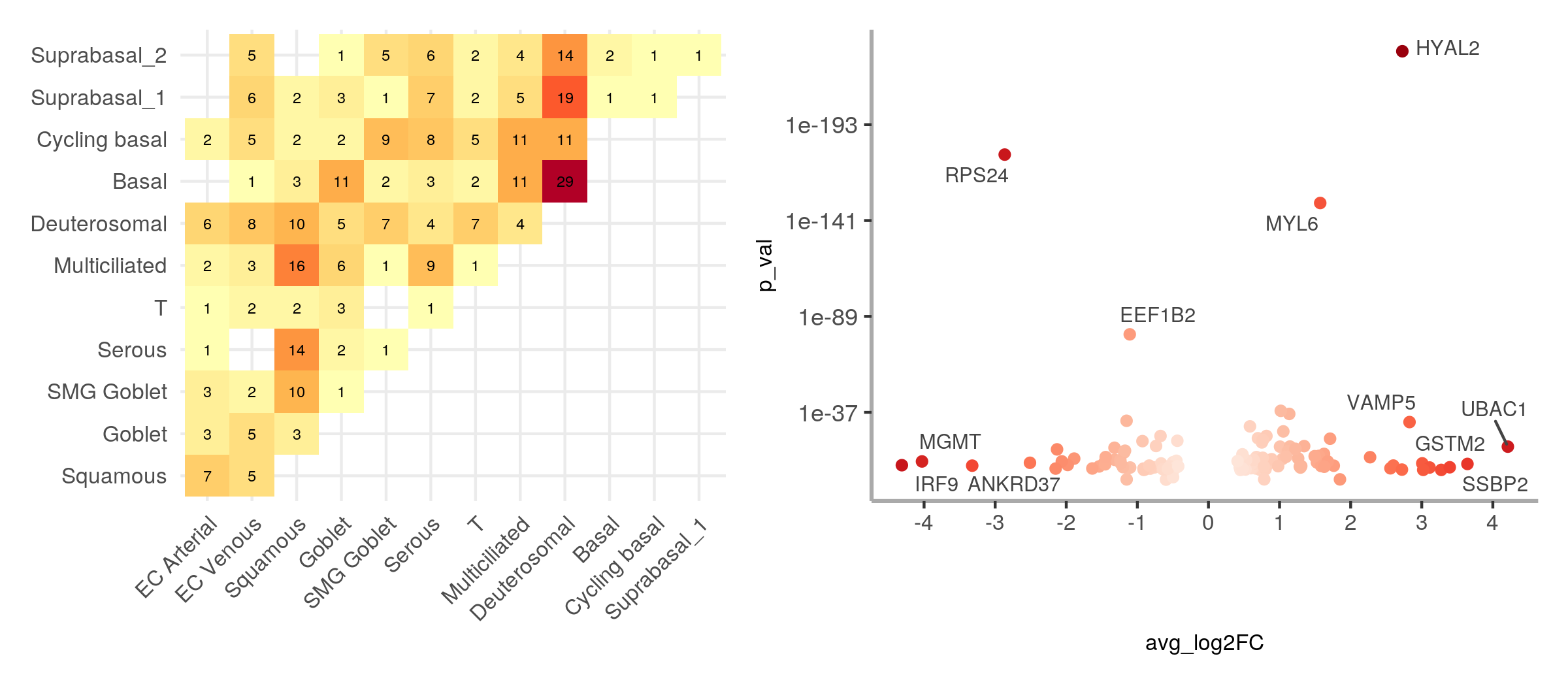
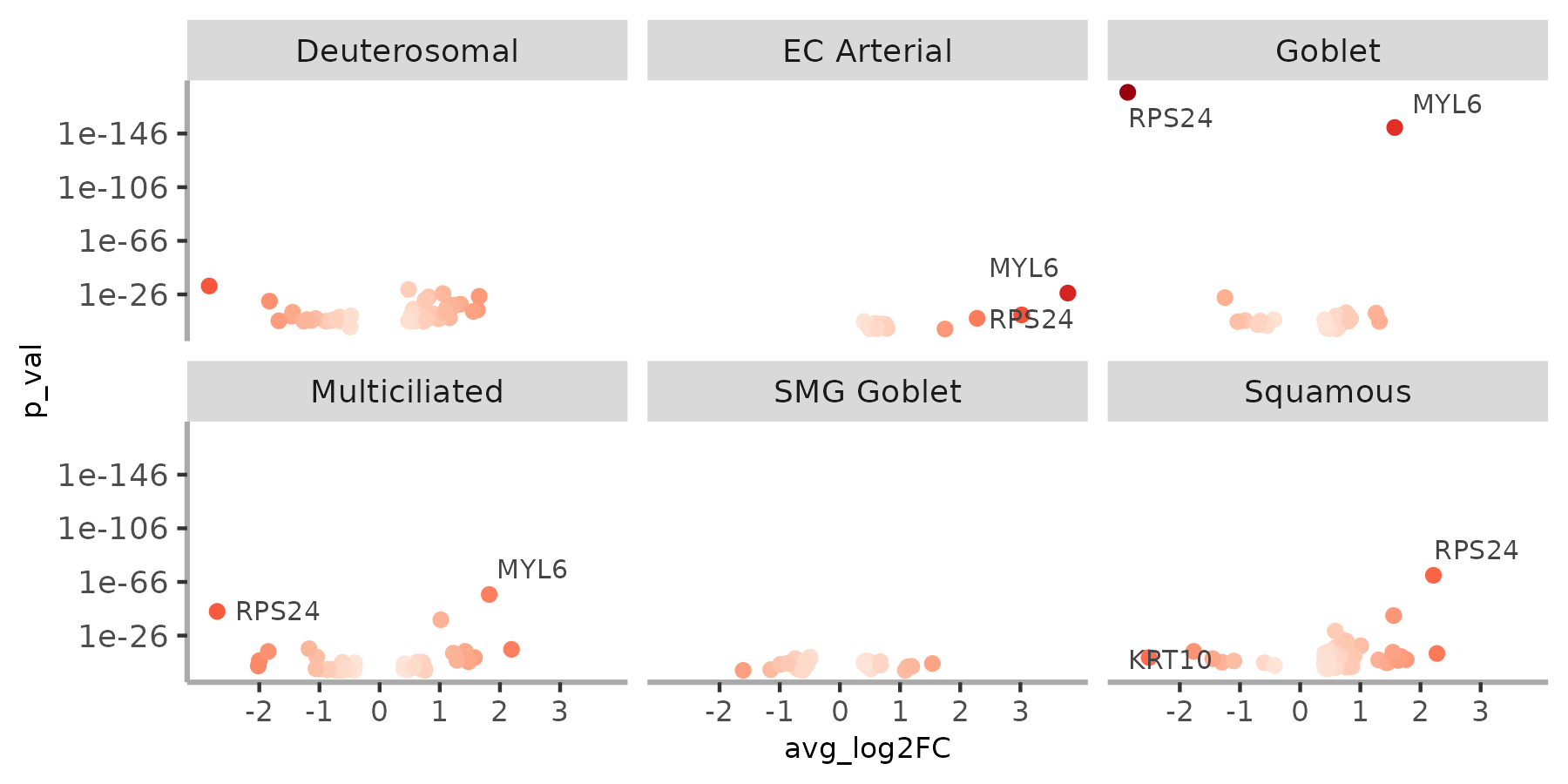
plot_marker_score() can
also plot individual volcano plots for each cluster analyzed with the
parameter facet=TRUE
plot_marker_score(adult, switch_markers, facet=TRUE, ncol=3)The method compute_switches() and gene_switch_table() compute
isoform switchs from a list of markrs and return a data.frame and an
html table respectively (only compatible with html output documents such
as Rmd reports)
switches <- compute_switches(switch_markers)
gene_switch_table(switch_markers)After identifying genes of interest, twere are two ways of drilling down and producing gene-level reports:
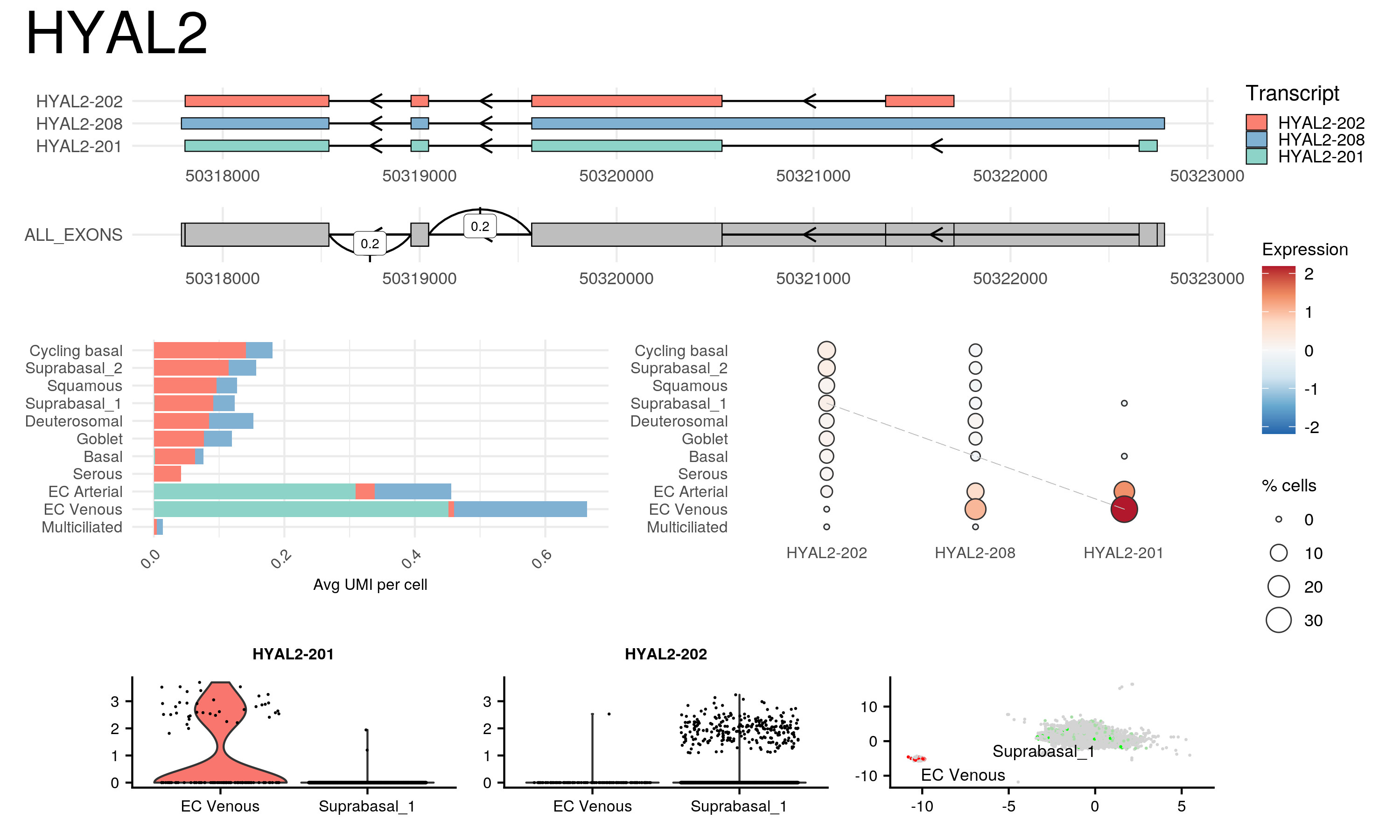
- The
isoswitch_report()method produces a compact plot of the gene. This function requires an extract from a gene annotation file and transcript metadata, documented here
isoswitch_report(seurat, "isoform", gene="HYAL2", marker_list=switch_markers, gtf_df, transcript_metadata) - The
render_html_gene_report()method renders an html version of the report