Authors: Hanwen Wang, Yifei Li, Zhenglin Zhang
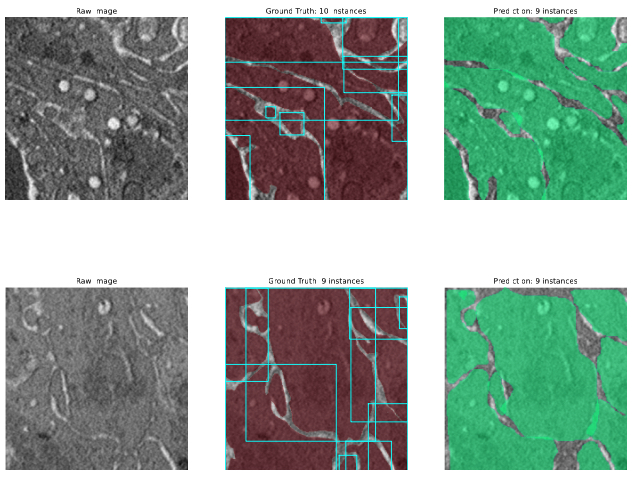
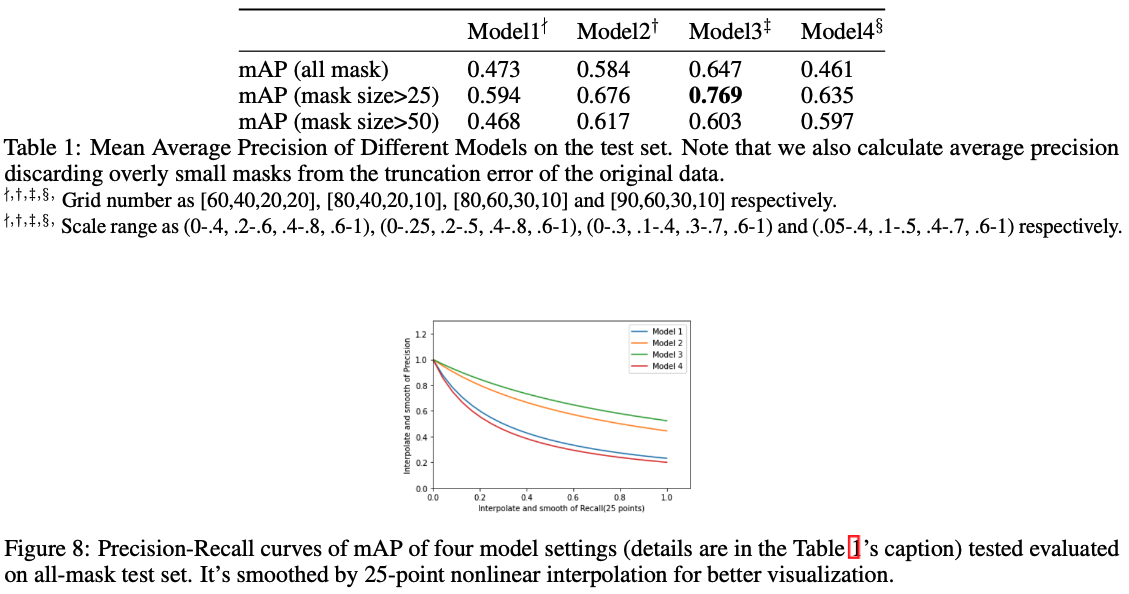
Abstract: In this project, we will explore the combination of traditional biomedical imaging dataset with state-of-the-art instance segmentation model. From moderated compressed dataset created with commercial software, we first use DBSCAN algorithm to convert them to standard data format that is compatible to the usage of data in computer vision field. We than modified the SOLO with a customized ResNet50 backbone for a binary classification problem, and searched a few hyperparameters to tune the model from COCO dataset to biomedical dataset. The average precision of the four sets of hyperparameters are 0.473, 0.584, 0.647 and 0.461. We also make a few attempts to add GRU for FPN output levels, but due to the extra complexity that it brings, none of them succeeds.
The report can be found here. However, due to the restricted accessibility of the original data, we're not allowed to share the image files.
- DBSCAN Algorithm - image conversion
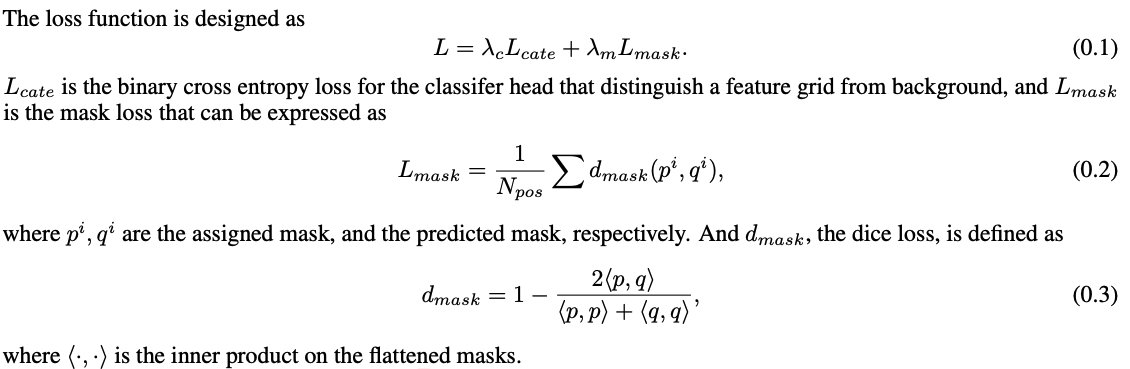
- Segmenting Objects by Locations (SOLO) - instance segmentation
- ResNet50 - FPN backbone #1
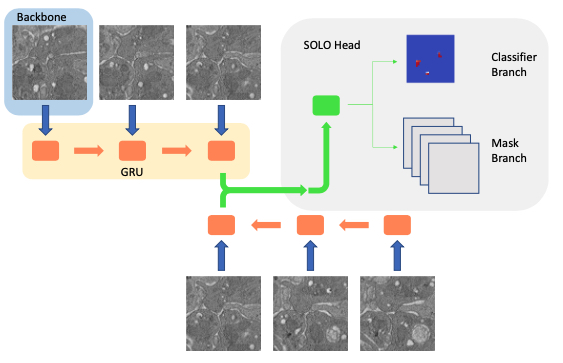
- Bi-GRU (z-axis) - FPN backbone #2
- Implement SOLO v2 since it’s more efficient on memory
- Try to use CenterNet as head or Swin Transformer as backbone