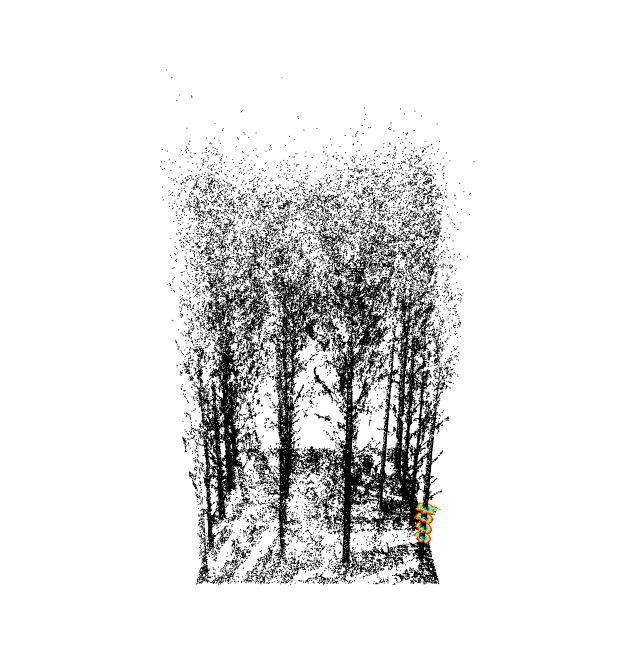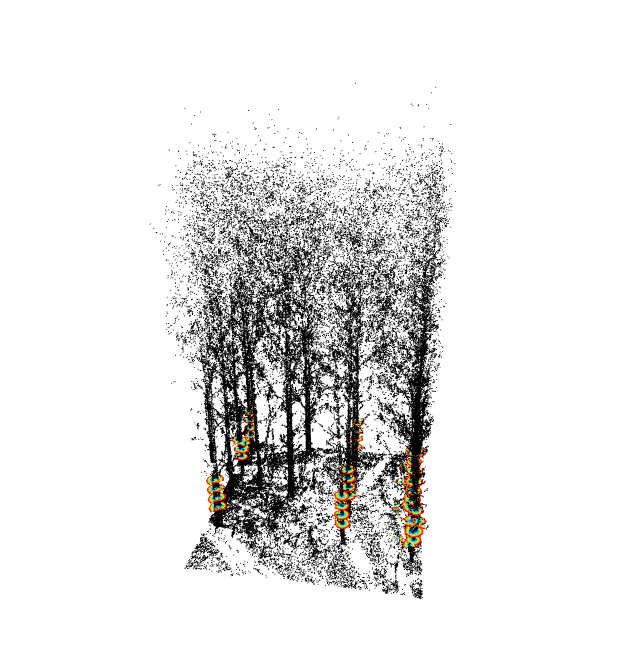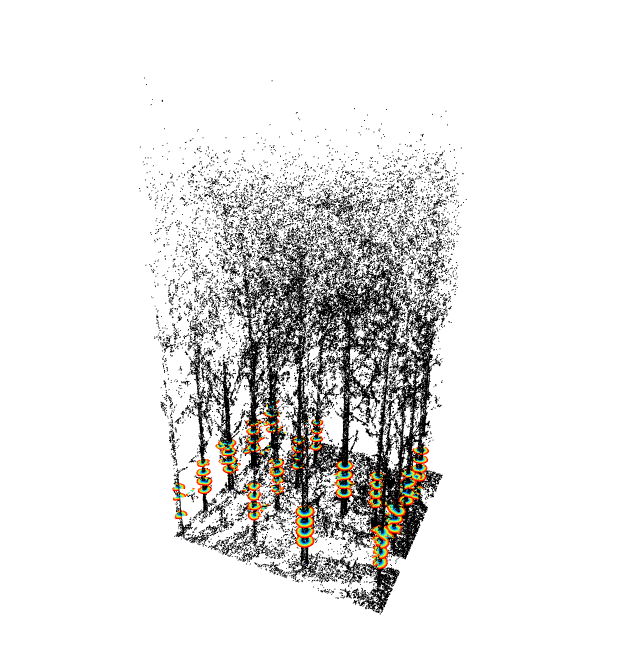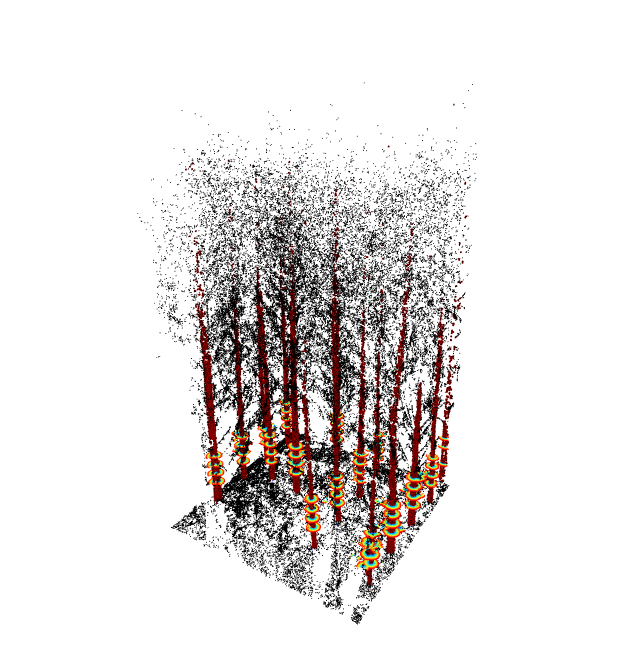High performance R functions for forest inventory based on Terrestrial Laser Scanning (but not only) point clouds.
This package is a refactor of the methods described in this paper.
The algorithms were rewritten in C++ and wrapped in R functions through Rcpp. The algorithms were reviewed and enhanced, new functionalities introduced and the rebuilt functions now work upon lidR's LAS objects infrastructure.
This is an ongoing project and new features will be introduced often. For any questions or comments please contact me through github. Suggestions, ideas and references of new algorithms are always welcome - as long as they fit into TreeLS' scope.
TreeLS v1.0 was released on CRAN as of March 2019. To install it from an official mirror, use: install.packages("TreeLS"). To install the most recent version, check out the Installation from source section below.
- March/2019:
TreeLSis finally available on CRAN and is now an official R package !!
- Tree detection at plot level
- Stem points detection at single tree and plot levels
- Stem segmentation at single tree and plot levels
lidRwrappers for writing TLS data with extra header fields- Eigen decomposition feature detection for trees and stems
- Tree modelling based on robust cylinder fitting
- 3D interactive point cloud manipulation
devtools: runinstall.packages('devtools', dependencies = TRUE)from the R console- Rcpp compiler:
- on Windows: install Rtools for your R version - make sure to add it to your system's path
- on Mac: install Xcode
- on Linux: be sure to have
r-base-devinstalled
On the R console, run:
devtools::install_github('tiagodc/TreeLS')
For anyone still interested in the old implementations of this library (fully developed in R, slow but suitable for research), you can still use it. In order to do it, uninstall any recent instances of TreeLS and reinstall the legacy version:
devtools::install_github('tiagodc/TreeLS', ref='old')
Not all features from the old package were reimplemented using Rcpp, but I'll get there.
Example of full processing pipe until stem segmentation for a forest plot:
library(TreeLS)
# open artificial sample file
file = system.file("extdata", "pine_plot.laz", package="TreeLS")
tls = readTLS(file)
# normalize the point cloud
tls = tlsNormalize(tls, keepGround = T)
plot(tls, color='Classification')
# extract the tree map from a thinned point cloud
thin = tlsSample(tls, voxelize(0.05))
map = treeMap(thin, map.hough(min_density = 0.03))
# visualize tree map in 2D and 3D
xymap = treePositions(map, plot = TRUE)
plot(map, color='Radii')
# classify stem points
tls = stemPoints(tls, map)
# extract measures
seg = stemSegmentation(tls, sgmt.ransac.circle(n = 15))
# view the results
tlsPlot(tls, seg)
tlsPlot(tls, seg, map)