This is a repository containing PyTorch code for PulseImpute: A Novel Benchmark Task for Physiological Signal Imputation. You can find our key visualizations from the paper as well as instructions for for testing pre-trained models in the paper and training your own models in the instructions below.
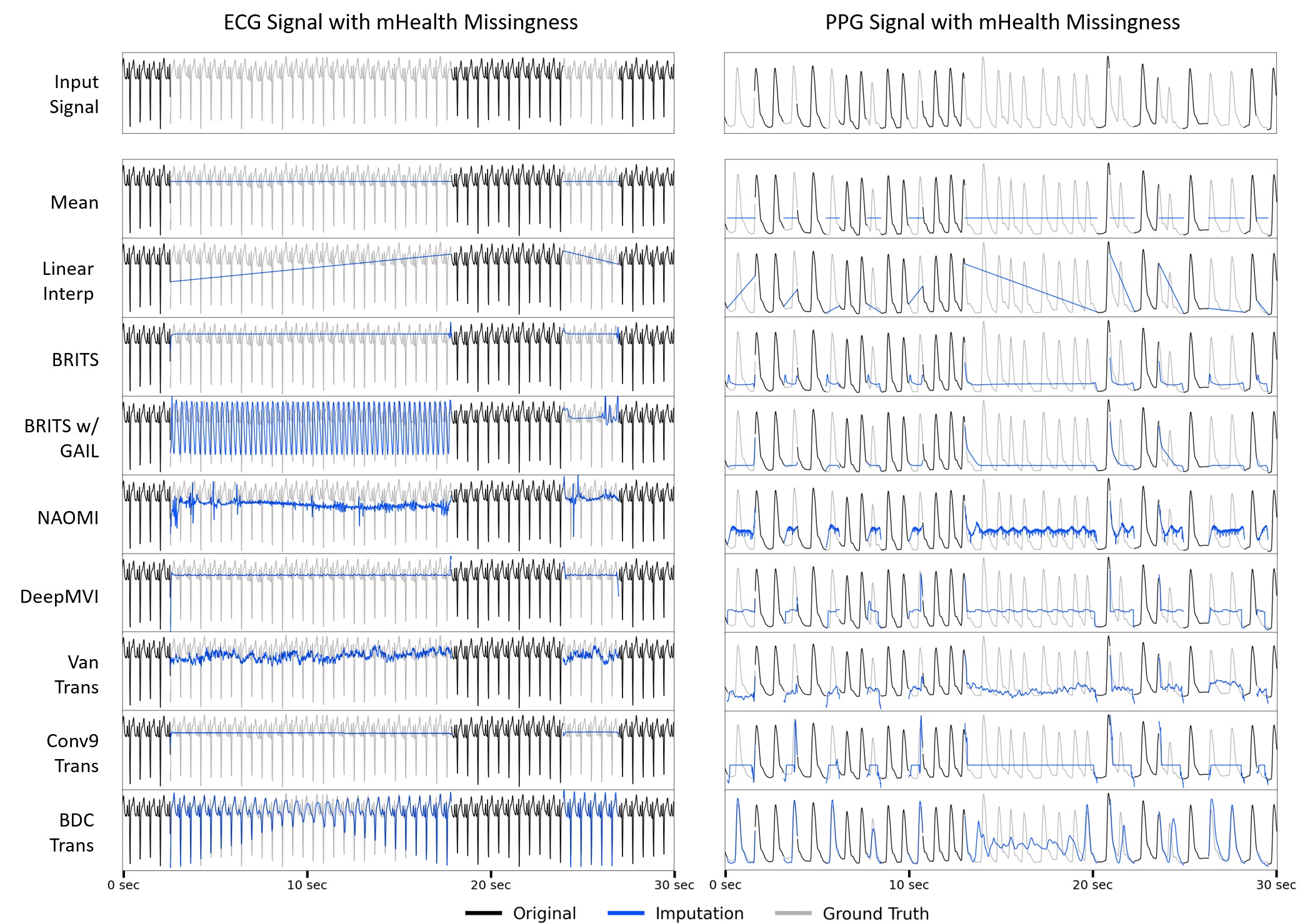
Visualization of imputation results from six baseline methods on representative ECG and PPG signals. The large temporal gaps which arise from real-world missingness patterns create substantial challenges for all methods. For example, methods such as BRITS and DeepMVI produce nearly constant outputs, while GAN-based approaches (BRITS w/ GAIL and NAOMI) hallucinate incorrect structures in incorrect locations. Our novel BDC Transformer architecture also struggles in the middle of long gaps.
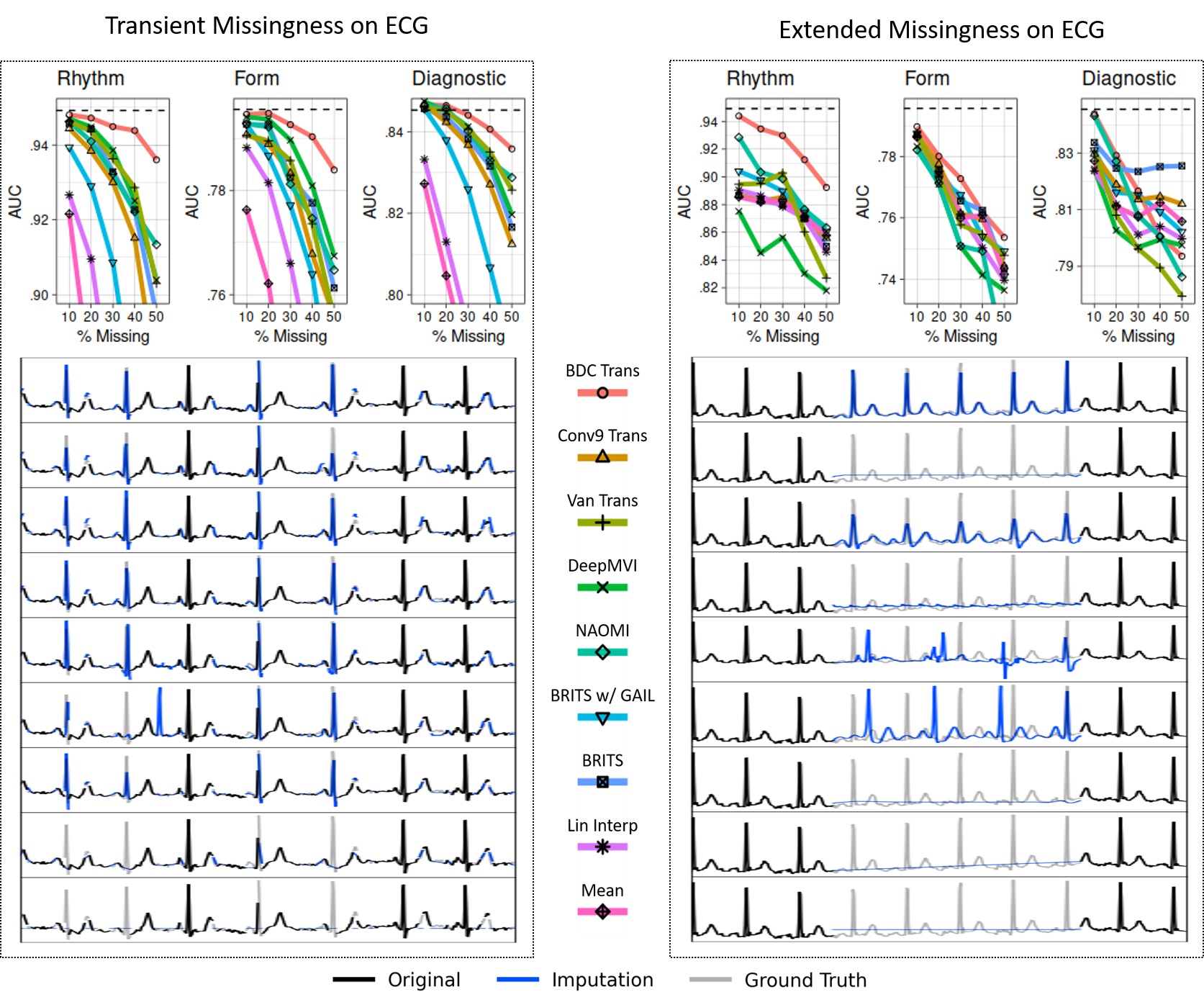
Cardiac Classification in ECG Results for Transient and Extended Loss on Rhythm, Form, and Diagnosis label groups. For each label group, a cardiac classifier was trained and tested on complete data (test performance illustrated by dashed line). The trained model was then evaluated on imputed test data (for five levels of missingness from 10% to 50%) produced by each baseline, yielding the AUC curves (top). Representative imputation results for the 30% missingness test case are plotted (below). The Extended Loss setting proved to be more challenging for all methods.
For this project we use miniconda to manage dependencies. After setting it up, you can install the pulseimpute environment:
conda env create -f pulseimpute.yml
conda activate pulseimpute
Download our extracted mHealth ECG/PPG missingness patterns and curated datasets (MIMIC-III ECG/PPG Waveform and PTB-XL) via the follwing bash-script:
./get_data.sh
This script downloads and extracts the ECG missingness patterns (missing_ecg_{train/val/test}.csv)), PPG missingness patterns (missing_ppg_{train/val/test}.csv)), MIMIC-III ECG data (mimic_ecg_{train/val/test}.npy)), MIMIC-III PPG data (mimic_ppg_{train/val/test}.npy)), and PTB-XL ECG data (ptbxl_ecg.npy)) from the data hosted here (469.8 MB compressed and 91.5 GB uncompressed). They can also be accessed via a persistent dereferenceable identifier at www.doi.org/10.5281/zenodo.7129965
Download our pre-trained checkpoints and imputed waveforms for each benchmarked models (BDC Transformer, Conv9 Transformer, Vanilla Transformer, DeepMVI, NAOMI, BRITS w/ GAIL, BRITS) for each PulseImpute task (ECG Heart Beat Detection, PPG Heart Beat Detection, ECG Cardiac Pathophysiology Classification) via the following bash script from the ckpts hosted here.
./get_ckpts.sh
Simply find the config of the model to be trained in their respective config file (e.g. configs/train_transformer_configs.py, configs/train_brits_configs.py, configs/train_naomi_configs.py) and in train_imp.py, set the config to that dictionary name to retrain each model from scratch.
python train_imp.py
Create the desired model class by following the example shown in models/tutorial/tutorial.py. The training and test methods for this tutorial model are in models/tutorial_model.py. Create a corresponding config entry in configs/train_tutorial_configs.py and run the training script train_imp.py with this config.
Simply find the config of the model to be tested in their respective config file (e.g. configs/test_transformer_configs.py, configs/test_brits_configs.py, configs/test_naomi_configs.py) and in train_imp.py, set the config to that dictionary name to run the imputation model on the test set.
python test_imp.py
We have created an interactive Google colab notebook that can be used to view the imputations of different models. No additional training or code running is required; the notebook can be run out of the box on a Jupyter environment.
Click play at the bottom left on the video below to watch a demo of the notebook:
visual_demo_v2.1.mp4
Explore the visualizations here!
If you use our work in your research, please cite
@article{xu2022pulseimpute,
title={PulseImpute: A Novel Benchmark Task for Pulsative Physiological Signal Imputation},
author={Xu, Maxwell and Moreno, Alexander and Nagesh, Supriya and Aydemir, Varol and Wetter, David and Kumar, Santosh and Rehg, James M},
journal={Advances in Neural Information Processing Systems},
volume={35},
pages={26874--26888},
year={2022}
}and if you have any further questions, please feel free to email me at maxxu@gatech.edu