Script to call L1 insertions and their methylation levels in bs-ATLAS-seq experiments.
If you use this script in your work, please cite:
Lanciano, S, et al. (2023). Comprehensive locus-specific L1 DNA methylation profiling reveals the epigenetic and transcriptional interplay between L1s and their integration sites. bioRxiv, 2023.01.03.522582; doi: https://doi.org/10.1101/2023.01.03.522582
[abstract] [full text]
Make sure to include the path to these programs in your $PATH variable by editing the appropriate file depending on your system (it could be ~/.bash_profile, ~/.bashrc, or ~/.profile)
- bash (tested with v4.2 and above, requires associated arrays)
- GNU coreutils (tested with v9.1)
- cutadapt (tested with v3.1)
- bowtie2 (tested with v2.4.1)
- bismark (version >= 0.22.0 which supports soft-clipping, tested with v0.22.1)
- samtools (tested with v1.3)
- Picard tools (tested with v2.19)
- bedtools (tested with v2.25.0)
- methpat (tested with v2.1.0)
- seqtk (tested with v1.3)
- GNU parallel (tested v20200922)
Note that <DOWNLOAD> is the name of the folder in which you have downloaded the repository.
- Download the project repository from GitHub:
wget 'https://github.com/retrogenomics/bs-ATLAS-seq/archive/refs/heads/main.zip' -O <DOWNLOAD>/main.zip
cd <DOWNLOAD>
unzip main.zip
mv bs-ATLAS-seq-main/ bs/
- Download a human reference genome (e.g. hg38):
mkdir -p bs/references/hg38
wget 'ftp://hgdownload.cse.ucsc.edu/goldenPath/hg38/bigZips/hg38.fa.gz' -O bs/references/hg38/hg38.fa.gz
gunzip bs/references/hg38/hg38.fa.gz
- Prepare repeatmasker file:
gunzip bs/annotations/hg38_rmsk_L1_repPercent.bed.gz
- Prepare Bismark human reference genome: Note that this can take several hours.
bismark_genome_preparation --verbose bs/references/hg38/
- Edit the
config_dir.shfile in thebs-ATLAS-seq/folder according to your configuration
We provide annotation files (genes, reference L1s, ENCODE black-list) for hg38. If you want to use another assembly, you can use the LiftOver tool to convert coordinates in your favorite assembly.
usage: bs-atlas-seq_calling.sh [options] -1 read1.fastq.gz -2 read2.fastq.gz -p prefix
options:
-h Print this help menu.
-v What version of bs-atlas-seq_calling are you using?
-o Output directory.
-n Total read number threshold to call an insertion [default=3]
-s Split read number threshold to call an insertion [default=2]
-u Subsampling of input fastq file (no=1; or indicate fraction of reads to consider, e.g. 0.01) [default=1]
-t Number of threads used for mapping [default=4]
-c Activate cleanup (deletion of temporary files) [default=off]
-d Configuration file [default=./config_dir.sh]
The pipeline generates a number of files which are described below:
| File name | Description |
|---|---|
<PREFIX>.bs-atlas-seq_calling.sh.script.sh |
A copy of the script used to generate the data. |
<PREFIX>.stats.txt |
A summary file with the number of reads, processed at each step, and the number of L1 detected. |
<PREFIX>.log |
A log file with the information displayed on the screen while the script is running. |
<PREFIX>.hg38.bam |
Alignment of the reads to the reference genome provided. |
<PREFIX>.hg38.bai |
Index file of the .bam file. |
<PREFIX>.all_L1.flanks_and_internal.hg38.bedGraph.gz |
BedGraph file with mCG levels. |
<PREFIX>.all_L1.hg38.bed |
Bed file containing all called L1 as well as their average mCG level as score (5th column). |
<PREFIX>.L1HS.hg38.bed |
Similar to <PREFIX>.all_L1.hg38.bed but filtered to keep only L1HS elements. |
<PREFIX>.all_L1.hg38.html |
An .html file displaying the single-molecule profiles of methylation for each L1 locus identified. Can be opened with a web browser (tested with Chrome). |
<PREFIX>.L1HS.hg38.html |
Similar to <PREFIX>.all_L1.hg38.html but filtered to keep only L1HS elements. |
The output directory will be ~/Downloads/test_220923 and output files will all be prefixed with test_IMR90.. Here we use the subsampling -u option to use only 1% of the reads for testing purposes.
./bs/scripts/bs-atlas-seq_calling.sh \
-1 ~/Lab/bioinfo/data/bs-atlas-seq/180525_NB501350_0027_AH37TJBGX7/IMR90_S6_R1.fastq.gz \
-2 ~/Lab/bioinfo/data/bs-atlas-seq/180525_NB501350_0027_AH37TJBGX7/IMR90_S6_R2.fastq.gz \
-o ~/Downloads/test_220923 \
-p test_IMR90 \
-n 3 -s 1 -t 6 -u 0.01 \
-d ~/Downloads/bs/config_dir.sh
.logfile
*********************************************************************************************************
[23-09-2022] [15:07:57] bs-ATLAS-seq analysis - Script v1.2.0
*********************************************************************************************************
Command line:
bs-atlas-seq_calling.sh -1 /Users/gcristof/Lab/bioinfo/data/bs-atlas-seq/180525_NB501350_0027_AH37TJBGX7/
IMR90_S6_R1.fastq.gz -2 /Users/gcristof/Lab/bioinfo/data/bs-atlas-seq/180525_NB501350_0027_AH37TJBGX7/IMR
90_S6_R2.fastq.gz -o /Users/gcristof/Downloads/test_220923 -p test_IMR90 -n 3 -s 1 -t 6 -u 0.01 -d /Users
/gcristof/Downloads/bs/config_dir.sh
*********************************************************************************************************
PE-sequencing data:
- R1: /Users/gcristof/Lab/bioinfo/data/bs-atlas-seq/180525_NB501350_0027_AH37TJBGX7/IMR90_S6_R1.f
astq.gz
- R2: /Users/gcristof/Lab/bioinfo/data/bs-atlas-seq/180525_NB501350_0027_AH37TJBGX7/IMR90_S6_R2.f
astq.gz
Ref genome:
- hg38: /Users/gcristof/Downloads/bs/references/hg38/hg38.fa
Sample:
- test_IMR90
*********************************************************************************************************
Steps:
1 - Data loading with subsampling to 0.01 fraction of sequences...Done
2 - Read trimming...Done
3 - Mapping R2 to L1...Done
4 - Mapping PE reads to hg38...Done
5 - Mapping discordant R1 reads to hg38...Done
6 - Identify split R1 reads...Done
7 - Merge all reads...Done
8 - Call reference L1 elements...Done
9 - Call non-reference L1 elements...Done
10 - Call CpG methylation for reference L1 elements...Done
11 - Call CpG methylation for non-reference L1 elements...[bam_merge] File '/Users/gcristof/Downloads/test_220923/temp/test_IMR90.R2toL1HS.nonref-L1HS.filtered.bam' exists. Please apply '-f' to overwrite. Abort.
Done
12 - Calculate methylation patterns and entropy for each L1 locus...Done
13 - Generate bedGraph files for CpG upstream of called L1 loci...Done
14 - Annotate identified L1 elements...Done
*********************************************************************************************************
Sample test_IMR90
Unprocessed reads 285697
Trimmed reads 187827
Mapped to L1 133817
Mapped to hg38 as PE 98357
Mapped to hg38 as SE 899
Total mapped to hg38 99256
Deduplicated mapped reads 65873
Called L1 unfiltered 4687
- including L1HS 388
- including nonref L1HS 21
Called L1 filtered 3910
- including L1HS 358
-- including nonref L1HS 21
L1 with methylation call 3877
- including L1HS 358
-- including nonref L1HS 21
*********************************************************************************************************
[23-09-2022] [15:15:10] Running time: 00:07:13 (hh:mm:ss)
*********************************************************************************************************
.stats.txtfile
Sample test_IMR90
Unprocessed reads 285697
Trimmed reads 187827
Mapped to L1 133817
Mapped to hg38 as PE 98357
Mapped to hg38 as SE 899
Total mapped to hg38 99256
Deduplicated mapped reads 65873
Called L1 unfiltered 4687
- including L1HS 388
- including nonref L1HS 21
Called L1 filtered 3910
- including L1HS 358
-- including nonref L1HS 21
L1 with methylation call 3877
- including L1HS 358
-- including nonref L1HS 21
.L1HS.hg38.bedfile
| chr | start | end | name | mCG_frac | strand | family | ref | read_count | mCG_count | CG_count | ME | closest_gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
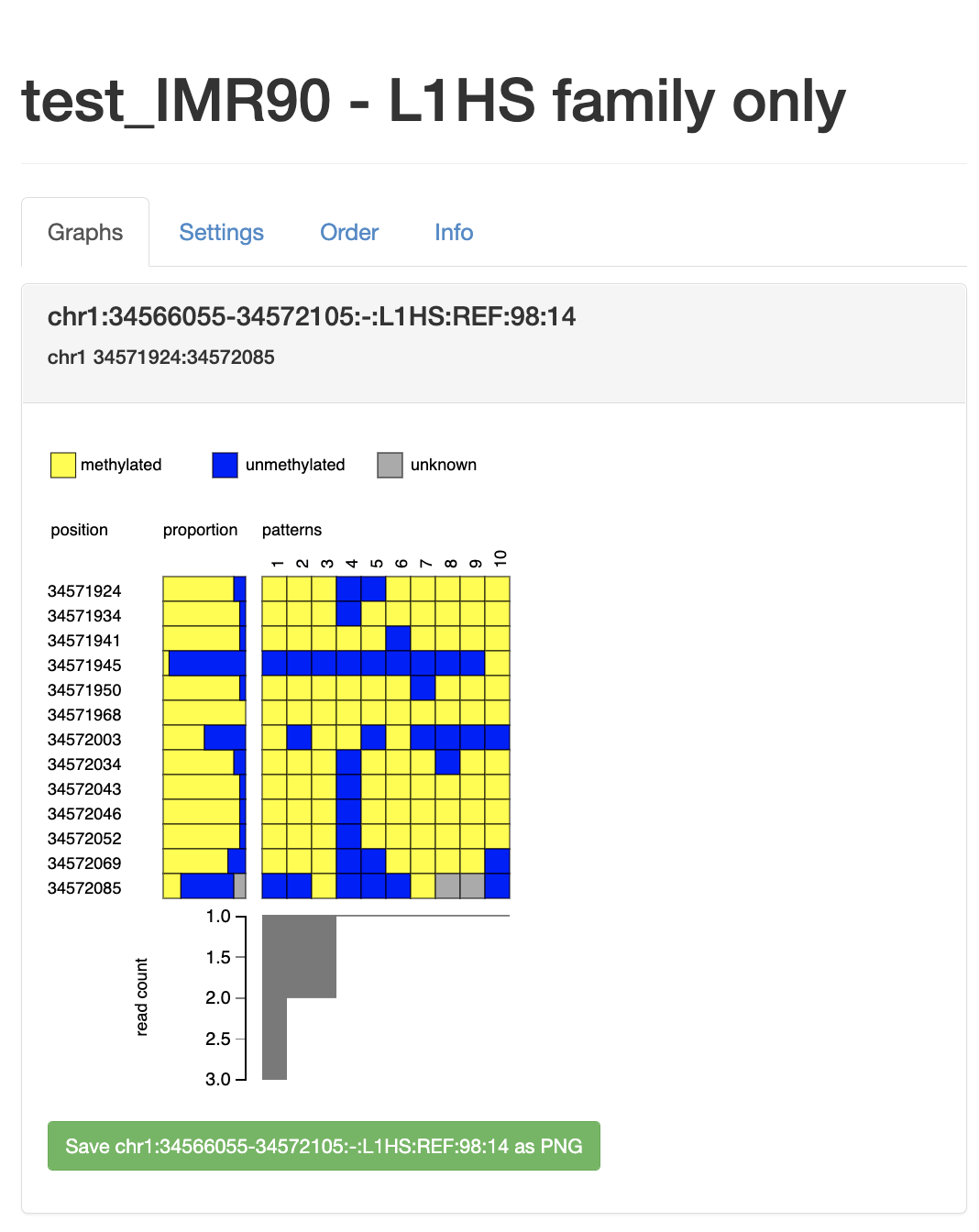
| chr1 | 34566055 | 34572105 | chr1:34566055-34572105:-:L1HS:REF:98:14 | 0.766667 | - | L1HS | REF | 14 | 138 | 180 | 0.247489 | . |
| chr1 | 67078891 | 67084915 | chr1:67078891-67084915:-:L1HS:REF:98:15 | 0.961722 | - | L1HS | REF | 15 | 201 | 209 | 0.155997 | C1orf141 |
| chr1 | 105315153 | 105315155 | chr1:105315153-105315155:-:L1HS:NONREF:100:11,4,3 | 0.876712 | - | L1HS | NONREF | 11 | 128 | 146 | 0.246943 | . |
| Field | Description |
|---|---|
| chr | chromosome |
| start | start coordinate |
| end | end coordinate |
| name | name of the L1 insertion built as chr:start-end:strand:family:ref:L1 length (% consensus):read_count [, softclipped read count, split read count for NONREF insertions] |
| mCG_frac | average L1 DNA methylation. |
| strand | L1 orientation |
| family | L1 family (e.g. L1HS, L1PA2, L1PA3, etc) |
| ref | REF for reference insertions; NONREF for non-reference insertions |
| read_count | number of reads supporting the L1 insertion |
| mCG_count | number of methylated CpGs |
| CG_count | total number of CpGs (both methylated and unmethylated) |
| ME | an estimate of methylation entropy (see Xie H. et al. Nucleic Acids Res 2011) |
| closest_gene | name of the closest gene in a 10 kb-window (a dot if none) |
.L1HS.hg38.htmlfile (can be opened with a web browser)