The original version of the MinHashed Atom-Pair fingerprint of radius 2 (MAP4) successfully combined circular substructure fingerprints and atom-pair fingerprints into a unified framework. This combination allowed for improved substructure perception and performance in small molecule benchmarks while retaining information about bond distances for molecular size and shape perception.
The present code expands the functionality of MAP4 to include encoding of stereochemistry into the fingerprint. Hereby, the CIP descriptors of chiral atoms are encoded into the fingerprint at the highest radius. This design choice allows to modulate the impact of stereochemistry on overall similarity, making it scale with increasing molecular size without disproportionally affecting structural similarity. The resulting fingerprint, MAP*, is calculated as follows:
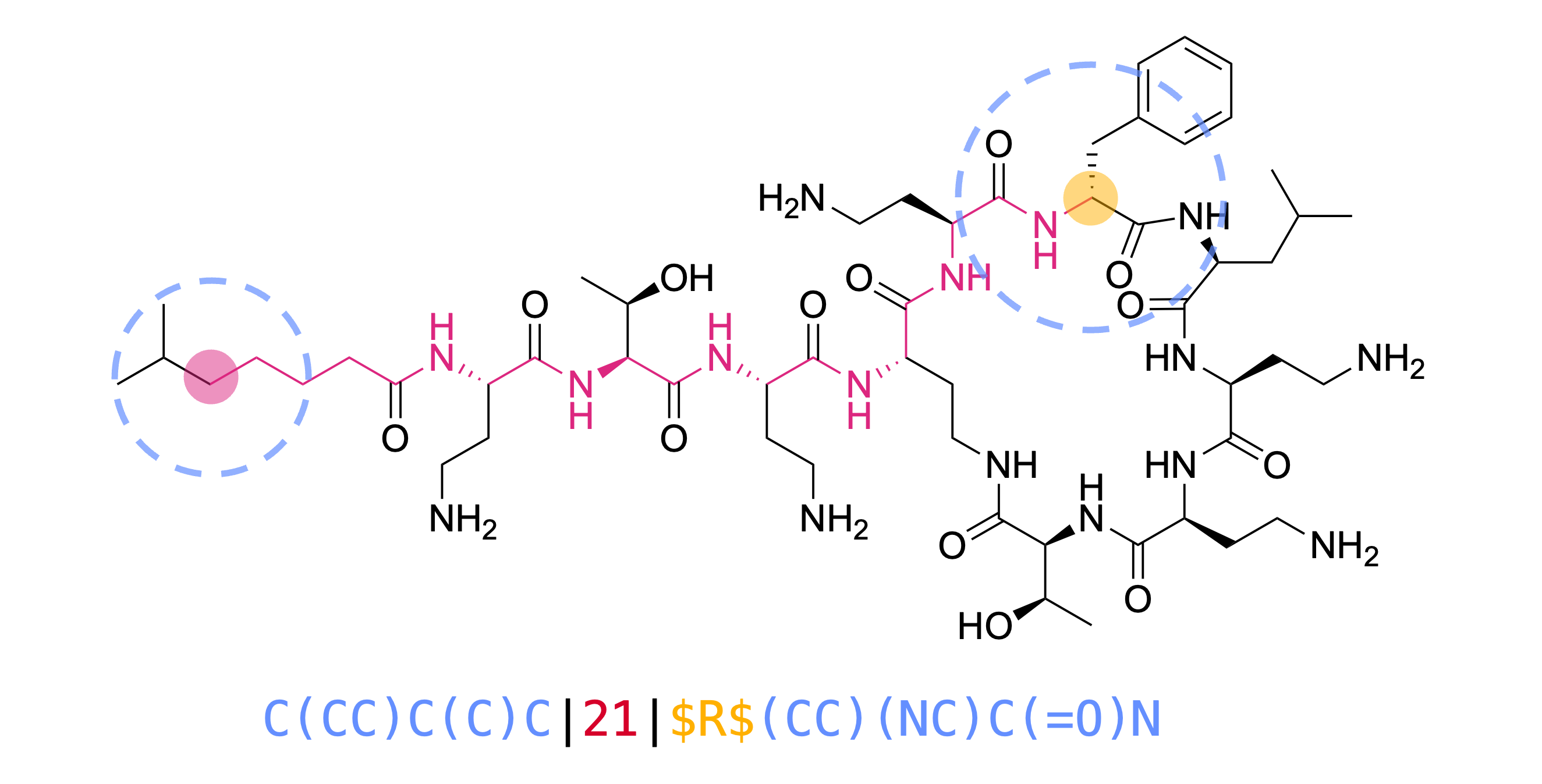
Fingerprint and shingle design. Every shingle contains two circular substructures (blue), the topological disance between the two substructures (red) and the CIP descriptor replacing the chiral atom (yellow).
The chiral version of the MinHashed Atom-Pair fingerprint (MAPC) was implemented in Python using RDKit following these steps:
-
At every non-hydrogen atom, extract all circular substructures up to the specified maximum radius as isomeric, canonical SMILES. Isomeric information (“@” and “@@” characters) is manually removed from the extracted SMILES, while the implicit E/Z-isomerism (“/”, and “\” characters) are maintained. Allene chirality and conformational chirality such as in biaryls or in helicenes are not considered, as they cannot be specified in the SMILES notation. Radius 0 is skipped.
-
At the specified maximum radius, whenever the central atom of a circular substructure is chiral, replace the first atom symbol in the extracted SMILES with its Cahn-Ingold-Prelog (CIP) descriptor bracketed by two “$” characters (
$CIP$ ). The CIP descriptor of the chiral atom is defined on the entire molecule, not on the extracted substructure. -
At each radius, generate shingles for all possible pairs of extracted substructures. Each shingle contains two substructures and their topological distance in following format: “substructure 1 | topological distance | substructure 2”.
-
MinHash the list of shingles to obtain a fixed sized vector. The MinHashing procedure is explained in detail in our previous publication.
Additional improvements to the original MAP4 code include:
-
Parallelization: The fingerprint calculation for a list of molecules is implemented in parallel using the multiprocessing library, which significantly reduces the calculation time for larger datasets.
-
Feature mapping: I included the option to map the hashes in the fingerprint to their respective shingles of origin. The idea is to enable the use of the fingerprint for explainable machine learning tasks. This function significantly increases the calculation time. It also gives different fingerprints than the non-mapped version as I had to change the minhashing function to make it work. Therefore, please be mindful when using this option and only use it when mapping is required. Also, if anyone has a better idea on how to implement this function, please let me know!
If you find any bugs, have suggestions for improvement or want to contribute to the code, please open a new issue and I will get back to you as soon as possible.
You will need following prerequisites:
To obtain a local copy of the project clone the GitHub repository as follows:
git clone https://github.com/reymond-group/mapchiral.gitCreate a new conda environment by running following command:
conda env create -f mapchiral.ymlActivate the environment:
conda activate mapchiralAnd finally you are ready to run the code from the cloned repository.
Alternatively, you can pip-install mapchiral on an existing Conda environment as follows:
conda activate my_environmentpip install mapchiralMAP* can be used for the quantitative comparison of molecules. The similarity between two molecules can calculated as the Jaccard similarity between their fingerprints using the function provided in the mapchiral package:
from rdkit import Chem
from mapchiral.mapchiral import encode, jaccard_similarity
molecule_1 = Chem.MolFromSmiles('C1CC(=O)NC(=O)[C@@H]1N2C(=O)C3=CC=CC=C3C2=O')
molecule_2 = Chem.MolFromSmiles('C1CC(=O)NC(=O)[C@H]1N2C(=O)C3=CC=CC=C3C2=O')
fingerprint_1 = encode(molecule_1, max_radius=2, n_permutations=2048, mapping=False)
fingerprint_2 = encode(molecule_2, max_radius=2, n_permutations=2048, mapping=False)
similarity = jaccard_similarity(fingerprint_1, fingerprint_2)
print(similarity)The mapchiral package also contains a function to calculate the fingerprints of a list of molecules simultaneously. This is especially useful for larger datasets as the calculation is parallelized and therefore much faster.
from rdkit import Chem
from mapchiral.mapchiral import encode_many
molecule1 = Chem.MolFromSmiles('C1CC(=O)NC(=O)[C@@H]1N2C(=O)C3=CC=CC=C3C2=O')
molecule2 = Chem.MolFromSmiles('C1CC(=O)NC(=O)[C@H]1N2C(=O)C3=CC=CC=C3C2=O')
molecules = [molecule1, molecule2]
fingerprints = encode_many(molecules, max_radius=2, n_permutations=2048, mapping=False, n_cpus=4)
print(fingerprints)Finally, the mapchiral package has an option (experimental) to map the hashes in the fingerprints to their original shingles. This is useful for explainable machine learning tasks. However, it significantly increases the calculation time. Mapping is also available for the "encode_many" function, where it returns a single dictionary containing all hashes present in the fingerprints and their respective shingles of origin.
from rdkit import Chem
from mapchiral.mapchiral import encode, jaccard_similarity
molecule_1 = Chem.MolFromSmiles('C1CC(=O)NC(=O)[C@@H]1N2C(=O)C3=CC=CC=C3C2=O')
fingerprint, hash_map = encode(molecule_1, max_radius=2, n_permutations=2048, mapping=True)
print(fingerprint)
print(hash_map)



