This repository contains key files and data for the article "Discovery of A Novel and Potent LCK Inhibitor for Leukemia Treatment via Deep Learning and Molecular Docking". Our work focused on finding new ligands for lymphocyte-specific protein tyrosine kinase (LCK).
- Data collection: For gathering the necessary data, we utilized a custom script named
1_Chembl_PDB_download.py (+ids.csv). This script is responsible for downloading active compound data from the ChEMBL database, filtering based on specific criteria, and preparing the data for subsequent analysis. It leverages libraries such as pandas, Chembl webresource client, and RDKit to handle chemical informatics tasks efficiently. The detailed procedure includes querying target compounds by Uniprot IDs, cleaning the retrieved SMILES strings, and downloading corresponding PDB files if available. - Data clustering: To analyze the collected data effectively, we employed a script called
1_cluster.py. This script reads an Excel file containing SMILES strings of molecules, calculates their molecular fingerprints using RDKit, and performs clustering based on the Tanimoto similarity of these fingerprints. The clustering is achieved through agglomerative hierarchical clustering, aiming to group compounds into 50 clusters. This step facilitates the identification of structurally similar compounds, enabling focused analysis and comparison within each cluster. - After completing the data collection and clustering steps, we have compiled the results into a consolidated dataset (
1_activates_from_chembl_clustered.xlsx). - Generation of Decoy Compounds and Statistical Analysis:
- Generation of Decoy Compounds: We leveraged our curated set of 50 active compounds as a blueprint to generate 2,900 decoy compounds using the DUD-E Database (DUD-E Database). This strategic generation ensures that the decoy compounds, while structurally reminiscent of the active set, exhibit distinct biological activities. This dichotomy is crucial for our validation framework, enhancing the robustness of our comparative study. The resulting datasets, encapsulated in
1_Decoys_smiles_from_DUDE.xlsxand1_activate_smiles_fromDUDE_50clustered.xlsx. - Molecular Property Analysis: To dissect the physicochemical essence of each compound within our dataset, active and decoy alike, we computed their molecular weight and partition coefficient (LogP). These computations were performed using the
Descriptors.MolWtandDescriptors.MolLogPfunctions from the RDKit library, respectively. This analytical step, executed through the1_logp.pyscript.
Our study also emphasizes the importance of visual representation in understanding the molecular and structural nuances of our selected compounds and proteins. To this end, we utilized two custom Python scripts for generating insightful plots:
- Script:
1_Verify.py - Purpose: This script creates a scatter plot comparing the Overall Quality Factor to VERIFY percentages for various protein structures. By plotting these metrics against each other and drawing a trend line, we gain insights into the correlation between structural quality and VERIFY scores. The plot also includes annotations for each structure’s PDB ID, providing a clear, at-a-glance understanding of the dataset's composition.
- Visualization: The resulting plot uses a color gradient to represent VERIFY percentages, enhancing the visual distinction between different structures based on their VERIFY scores.
- Script:
1_protein_donuts.py - Purpose: To visualize the distribution of amino acid residues in various regions of interest across the selected proteins, we employ donut plots. These plots categorize residues into most favoured regions, additional allowed regions, generously allowed regions, and disallowed regions, based on their Ramachandran plot positions.
- Visualization: Each protein is represented by a donut plot, with different colors indicating the distribution percentages of residues in the specified categories. The plots offer an intuitive way to assess the structural integrity and stereochemical quality of the proteins.
PDB structures and the assessments carried out using PyRAMA and SAVES v6.0, are comprehensively archived in 1_PDB.zip.
For a comprehensive assessment of our docking models, we utilized a variety of tools, each chosen for its specific strengths in simulating molecular interactions. Detailed parameters and methodologies are elaborated in our manuscript. Below is a summary of the tools used, along with their setup and execution protocols.
- Installation and Usage: Follow the official guidelines provided by AutoDock-GPU.
- Execution Scripts:
2_adgpu.shand2_adgpuanalysis.share utilized to run the docking simulations and analyze the results, respectively.
- Installation and Usage: Adhere to the official instructions available at AutoDock-Vina.
- Execution Script:
2_vina.shis used for running docking simulations.
- Installation and Usage: Installation and operation should conform to the instructions on the PLANET GitHub page.
- Installation and Usage: Guidelines are provided on the LeDock website. The script
2_ledock.shis employed for docking processes.
- Installation and Usage: Follow the official Schrödinger manuals for the 2021-4 version, accessible here.
Note: The molecular docking, MD simulation, and structural analysis conducted in this study were facilitated by the high-performance computing cluster platform at the School of Biotechnology, Jiangnan University (Schrodinger2021-4).
Furthermore, the outcome of our docking validation, including docking scores and related data, is comprehensively documented in the '2_Validation_dockingscore.xlsx' file.
In addition to the docking scores analysis, we employed the 2_AUCROC.py script for the statistical analysis of our results, specifically for calculating the Area Under the Receiver Operating Characteristic (AUC-ROC) curves and conducting t-tests. This further quantifies the predictive accuracy and statistical significance of our docking model's performance.
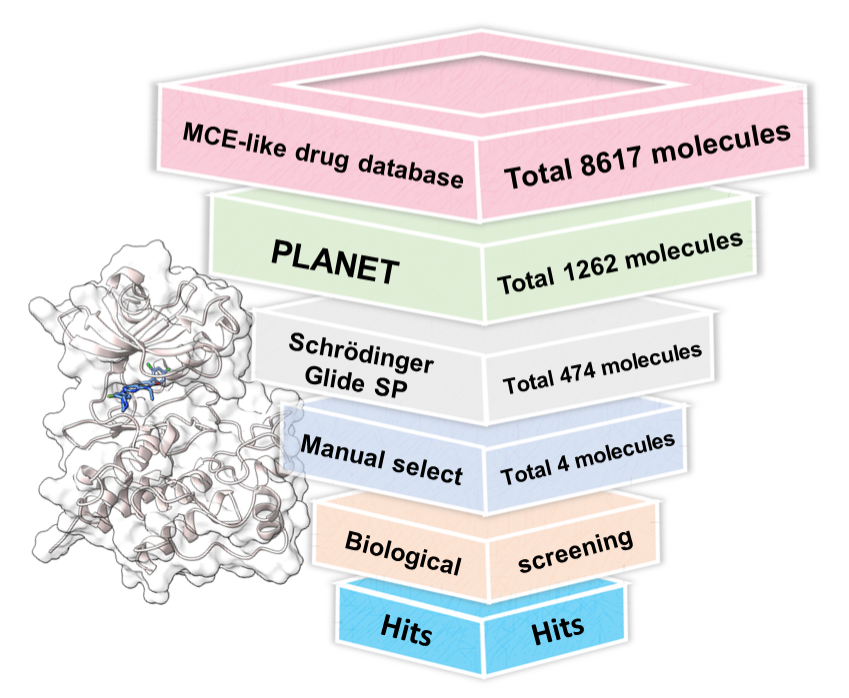
Our virtual screening workflow leverages the capabilities of PLANET and Schrödinger Glide SP to identify promising molecular candidates. For detailed parameters and methodologies, we refer readers to our manuscript. This section outlines key components of our workflow, including molecule filtration and similarity distribution, and the outcome of our screening process.
- Filtered Molecules: The filtration of molecules was conducted using the
3_filtered_molecules.pyscript. - Distribution of Maximum Similarity: To assess the chemical diversity and similarity of our filtered molecules, we utilized the
3_similarity.pyscript. This analysis allowed us to understand the distribution of molecular similarities within our dataset, highlighting the range of chemical spaces covered by our candidates.
The culmination of our virtual screening process identified the top 474 molecules, which have been documented in the 3_474_compounds.sdf file. In addition, the docking scores, which provide a quantitative measure of the binding affinities between these molecules and the target protein, are meticulously detailed in the 3_474_docking_score.csv file.
-
Script Name: Protocol provided by Thibault Tubiana's protocolGromacs Developed by: Thibault Tubiana, PhD
-
Purpose This script employs an integrated tool developed by Thibault Tubiana, PhD, for conducting Molecular Dynamics (MD) simulations. It provides a convenient and efficient way to set up, run, and analyze MD simulations using GROMACS. We have conducted a simulation lasting 500 nanoseconds, with other specific parameters detailed in the manuscript.
-
File Management The Results File:
4_MDresults.zip
-
Source: gmx_mmpbsa.bsh git from Jerkwin's gmxtools
-
Tools: APBS-3.0.0.Linux available at APBS GitHub
-
Purpose The script
gmx_mmpbsa.bshis utilized for calculating MM-PBSA binding energies and performing energy decomposition for GROMACS trajectories. It automates the Molecular Mechanics (MM) calculations and uses the Adaptive Poisson-Boltzmann Solver (APBS) program for the Poisson-Boltzmann Surface Area (PBSA) calculations. This approach extracts necessary atomic parameters such as charges, radii, and Lennard-Jones (LJ) parameters from GROMACS.tprfiles via thegmx dumpcommand, ensuring compatibility across different versions of GROMACS. The script also supports APBS across its versions as long as there's no change in its input file format. -
File Management The Results File:
4_MMPBSA.zip