A collection of tools and scripts which are useful for maipulating XRD data and performing CMWP
- src/xrd_tools.py (library containing useful general XRD routines)
- getReflections: makes a list of peak hkl and position for given wavelength and crystal structure
- src/cmwp_tools.py (library contining useful commands specific to CMWP)
- load_tifs: Load images from files using fabio
- integrate_join: Integrate images from two detectors using pyFAI and join together
- calculateLatticeParams: Uses minimisation of an objective function based on the Hull-Davey formula to calculate lattice paramaters
- dislocationTypeCalc: Calculation of dislocation type from given
a1anda2values for a hcp crystal - extractDataDir: Extract physical paramaters from all CMWP solution files in a directory
- getBaseline: Produces a background spline for given 2theta positions
- getPeaks: Finds peak positions and intensity in a pattern, given approximate 2theta positions
- data/srim.txt (SRIM data for 2 MeV proton irradiation of Zr - 1dpa at 60% depth)
- data/ellipticities.txt (Chk0al, alaL and a2aL for given ellipticity - used for dislocation loop type calculation)
- data/test.xy (Test XRD xy data)
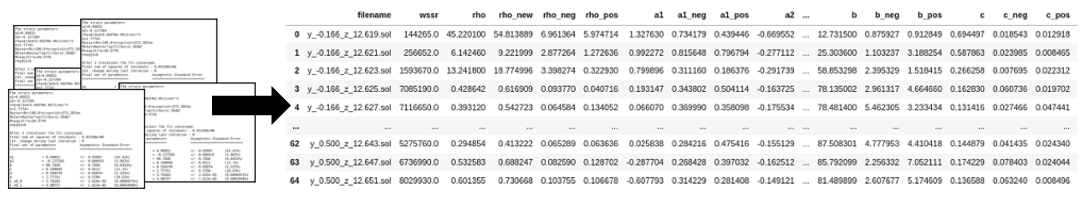
- Extract_data.ipynb (Data extraction from a folder of .sol files and calculation of a/c loop fraction)
- Extract_data-new.ipynb (Data extraction from a folder of .sol files for Wilkens function method)
- Zr_DESY_2021.ipynb (Integration of data from DESY experiment and production of .bg-spline.dat and .peak-index.dat)
- Zr_batch.ipynb (Production of .bg-spline.dat and .peak-index.dat for pre-integrated data)
- Label_reflections.ipynb (Load XRD data and label Zr/ZrH/SPP peaks)
- Make_CMWP_intrumental.ipynb - Generates an instrumental folder for CMWP from a LaB6 pattern