This repository is the official implementation of Correspondence Learning via Linearly-invariant Embedding
This code was written by Marie-Julie Rakotosaona and Riccardo Marin.
NEWS: A PyTorch implementation of the method can be found here
- CUDA and CuDNN
- Python 2.7
- Tensorflow 1.15
You can download pretrained models here:
Download pretrained models from code:
cd pretrained_models
python download_models.pyDownload dataset from code:
cd data
python download_data.pyTo train the basis model:
python src/train_basis.py --log_dir "path to basis model directory"
To train the transformation model:
python src/train_descriptors.py --log_dir "path to transformation model directory" --model_path "path to basis model"
To evaluate a model, you can use the "test.py" file. For example, to evaluate the pre-trained model on the Noise dataset:
Run pretrained model on FAUST testset:
python ./src/test.py --i FAUST_noise_0.01 -n log_model --epoch=4427
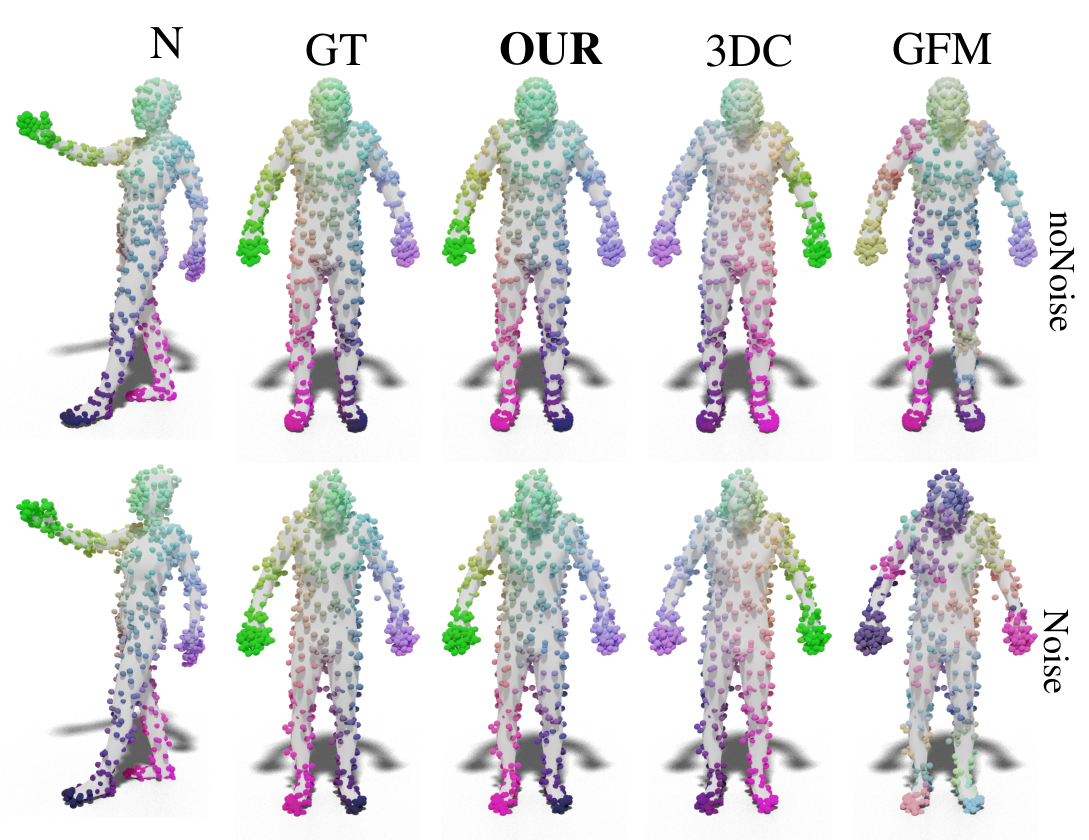
The evaluation of the correspondence for point clouds generated from the FAUST dataset without or with additional noise as mean error, with cumulative curves and a qualitative example.
| Method | No noise | Noise |
|---|---|---|
| our | 5.4e-2 | 6.6e-2 |
| GFM | 2.9e-1 | 3.4e-1 |
| Uni20 | 7.5e-2 | 8.5e-2 |
| Uni60 | 6.9e-2 | 8.1e-2 |
| 3DC | 7.0e-2 | 7.3e-2 |
| FMAP | 1.3e-1 | 1.4e-1 |
| FMAP+ZOO | 1.1e-1 | 1.3e-1 |
| GFM+ZOO | 3.1e-1 | 3.8e-1 |
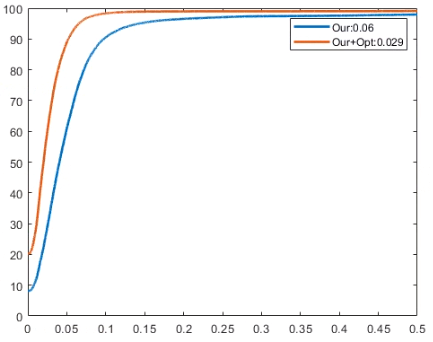
To replicate our evaluation, you can run the "evaluation.m" in Matlab. It is already configurated to run on the output of pre-trained model. In the paper we computed the geodesic distance matrices using an old pre-compiled .mex file for fastmarch algorithm. For the moment (and for the sake of semplicity), in this git we attached a pre-computed geodesic distance matrix used for all the pairs. It is an approximation, but this will let you to run the evaluation. This should return to you a plot like this:
If you use our work, please cite our paper.
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. For any commercial uses or derivatives, please contact us.