This is a public data set for analyzing how a gut microbiome is affected by drinking kombucha. You are welcome to study this as much as you like, provided you give your feedback. Contact me on Twitter @sprague or update the Wiki associated with this repository.
Update 3/21/2017: I just added some new ZIBR statistical analysis. It's not working yet.
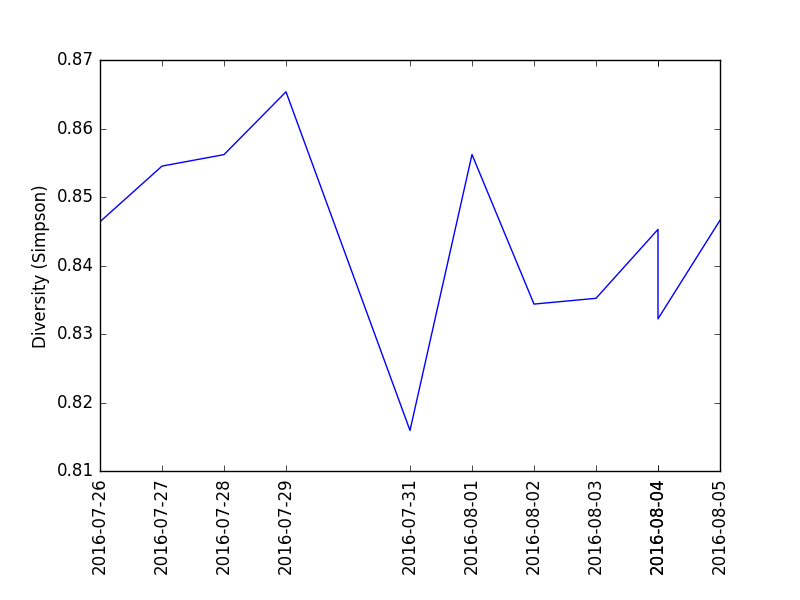
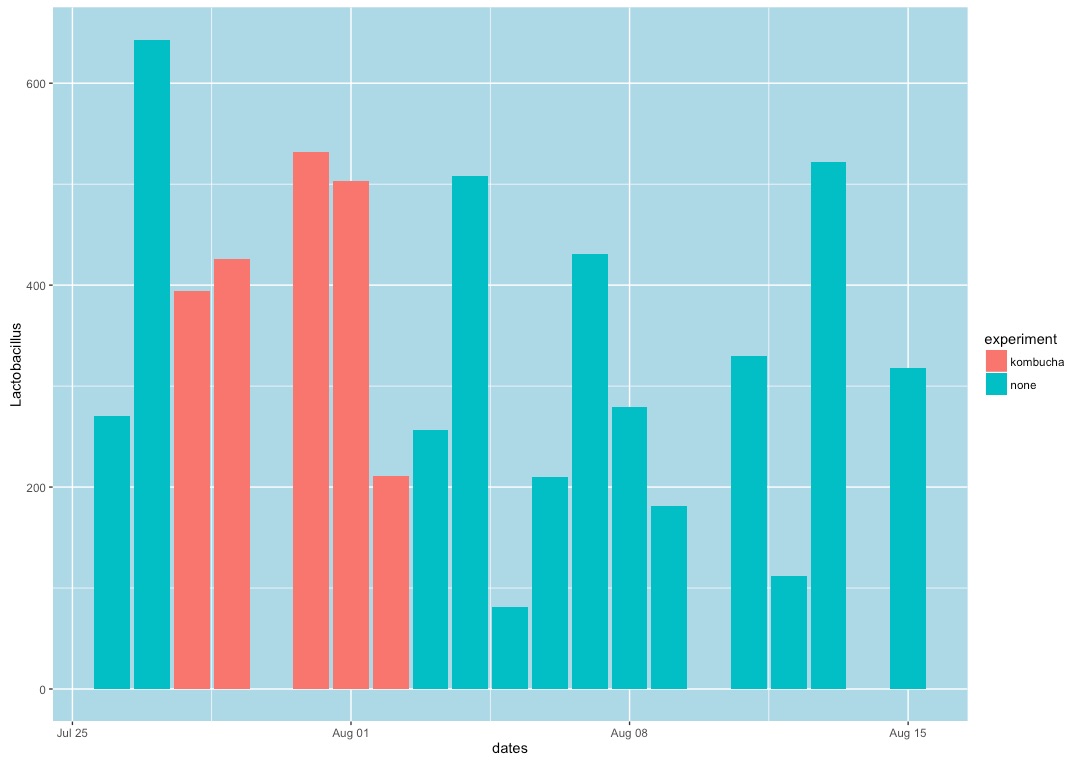
For seven days, from July 27 to August 2, 2016, I drank 48 ounces per day of commercially-purchased kombucha. I tested my gut microbiome with a uBiome Explorer kit each day, as well as my mouth and skin microbiome at intervals during the experiment.
The results of each uBiome Explorer test are in the data directory in a series of JSON-formatted files.
I am a middle-aged male in excellent health. I am 6 feet tall and weigh 160 pounds, take no prescription drugs, have never smoked tobacco, and live a reasonably active lifestyle. My appendix was removed as a child.
This data set also includes a complete list of all the foods I consumed between July 26th (the day before the experiment) and August 5th (several days afterwards). As you will see, I am an omnivore who eats a variety of foods, including coffee, dairy, wheat, meat, fish, and some alcohol.
If you have the uBiome open source python library, the enclosed Python script kombucha-analysis.py will read the uBiome JSON results files into a variable for further analysis, and plot the diversity like this:
You can do some interesting statistical analysis on this data if you use the R programming environment. The sample script kombucha.R will pull all the data into an R environment and draw the following chart:
There is much more analysis that can be done with this data. Some of the ideas you might try are:
- Study correlations among the taxa. Which ones are correlated, and which are not?
- Which taxa appeared and/or disappeared during the experiment?
- Is there a relationship between the microbes known to be present in kombucha and those in any of the gut results?
- How do these results compare to you when you drink kombucha?
Please study as much as you like, and let me know what you find!
Follow @sprague<script async src="//platform.twitter.com/widgets.js" charset="utf-8"></script>