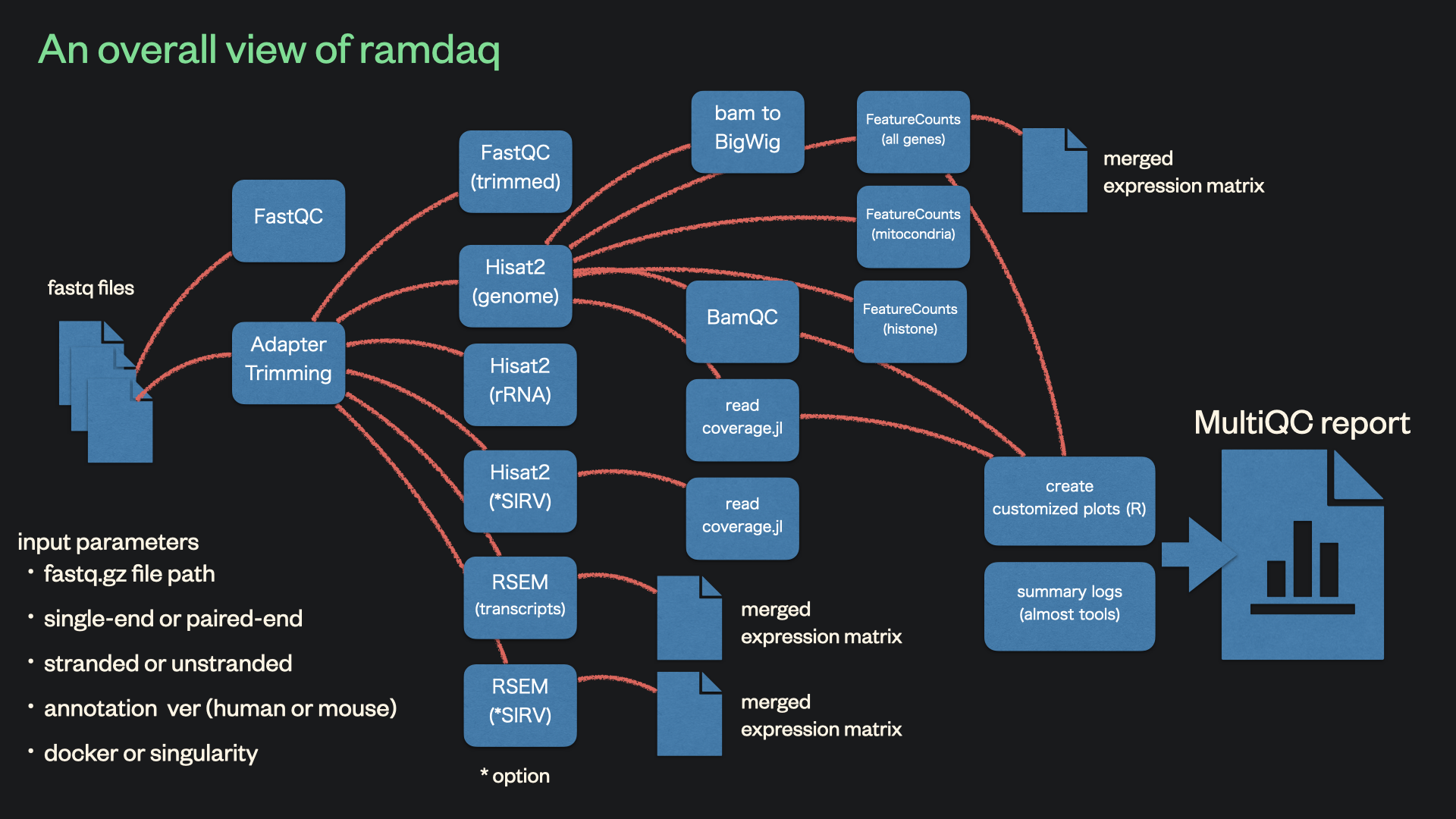
This pipeline analyses data from full-length single-cell RNA sequencing (scRNA-seq) methods.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker containers making installation trivial and results highly reproducible.
- Read QC (FastQC)
- Adapter and quality trimming (FastqMcf)
- Trimmed read QC (FastQC)
- Sort and index alignments (Hisat2 and SAMtools)
- Quantification of gene-level and transcript-level expression (RSEM)
- Generation of BigWig (coverage) files (bam2wig)
- Mapping/alginment QC:
- RSeQC
- readcoverage.jl
- Quantification of gene-level expression (featureCounts)
- Quantification of rRNA reads (HISAT2 and SAMtools)
- Alignment and quantification of SIRV reads (HISAT2, SAMtools, and RSEM) (optional)
- HTML QC report for raw read, alignment, gene biotype, sample similarity, and strand-specificity checks (MultiQC, R)
i. Install nextflow
ii. Install either Docker or Singularity for full pipeline reproducibility (see docs). Note that ramdaq does not support conda.
iii. Download the pipeline automatically and test it on a minimal dataset with a single command
1. Example of test using Docker
nextflow run rikenbit/ramdaq -profile test,docker2. Example of test using Singularity
nextflow run rikenbit/ramdaq -profile test,singularityiv. Start running your own analysis!
iv-i. You can run ramdaq without donwloading reference annotation data.
nextflow run rikenbit/ramdaq -profile <docker/singularity> --reads '*_R{1,2}.fastq.gz' --genome GRCh38_v37iv-i. You can also run ramdaq by specifying local paths to reference annotation (See 'Using provided reference genome and annotations').
nextflow run rikenbit/ramdaq -profile <docker/singularity> --reads '*_R{1,2}.fastq.gz' --genome GRCh38_v37 --local_annot_dir <The directory path where the reference genome and annotations are placed>See usage docs for all of the available options when running the pipeline.
To download or update ramdaq, run nextflow pull:
nextflow pull rikenbit/ramdaqTo check the available versions, run nextflow info:
nextflow info rikenbit/ramdaqThe above command will return the message like this (* master (default) indicates that the latest version will be used when you execute nextflow run rikenbit/ramdaq ...):
$ nextflow info rikenbit/ramdaq
project name: rikenbit/ramdaq
repository : https://github.com/rikenbit/ramdaq
local path : /Users/haruka/.nextflow/assets/rikenbit/ramdaq
main script : main.nf
description : This pipeline analyses data from full-length single-cell RNA sequencing (scRNA-seq) methods.
author : Mika Yoshimura and Haruka Ozaki
revisions :
* master (default)
dev
1.0 [t]
1.1 [t]To use versions other than the latest version, use -r to set the version name as follows:
nextflow run rikenbit/ramdaq -r 1.1 ...The ramdaq pipeline comes with documentation about the pipeline, found in the docs/ directory:
- Installation
- Pipeline configuration
- Running the pipeline
- Usage
- Examples
- Using test data
- Using bcl2fastq
- If you need to use BCL files produced by Illumina sequencing machines, execute ramdaq_bcl2fastq.
- bcl2fastq is conversion software, which can be used to demultiplex data and convert BCL files to FASTQ file formats for downstream analysis.
- Please see the README of ramdaq_bcl2fastq for details.
- Using provided reference genome and annotations
- the current version supports human (GRCh38) and mouse (GRCm38).
- Using ramdaq on the NIG Supercomputer System
- Output and how to interpret the results
- Troubleshooting
ramdaq is written and maintained by Mika Yoshimura and Haruka Ozaki in the collaboration of Laboratory for Bioinformatics Research, RIKEN Center for Biosystems Dynamics Research and Bioinformatics Laboratory, Faculty of Medicine, University of Tsukuba.
ramdaq was originally developed based on the nf-core template.