Codes for data processing and figure generation
install.packages(c("progess","plyr","ggplot2"))
import numpy
import matplotlib
import seaborn
import scipy
import pybedtools
import pysam
import json
import pandas
import re
import collections
import itertools
import tables
- Download and store the DNase file using the meta file information.
Rscript 1-a-download-DNase-ENCODE.R
- Rename DNase files
Rscript 1-b-recode-DNase.R
- Download hg19 chromosome length file
Rscript 1-c-hg19-information.R
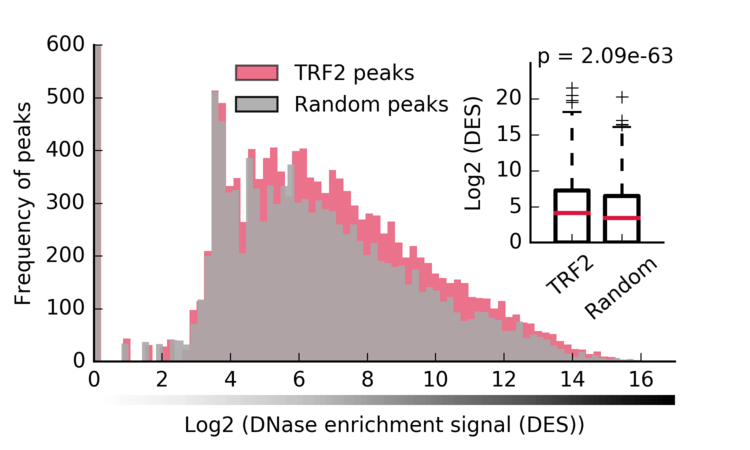
- DES in TRF2 vs Random
python 2-b-Figure-2A.py
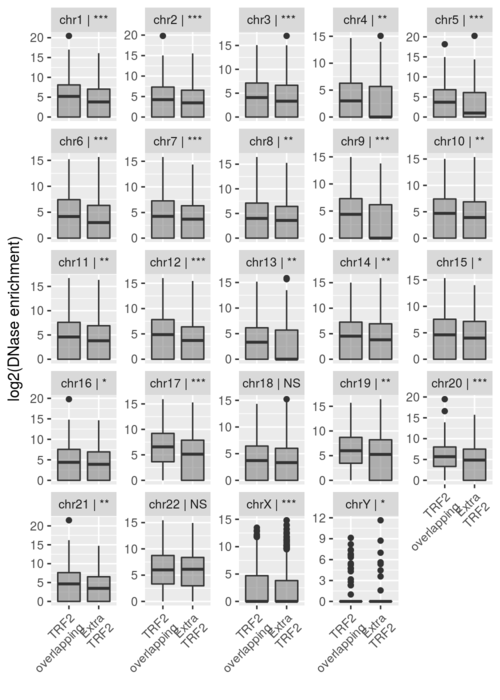
- DES per chromosome comparison
Rscript 2-c-Suppl-Figure-2.R
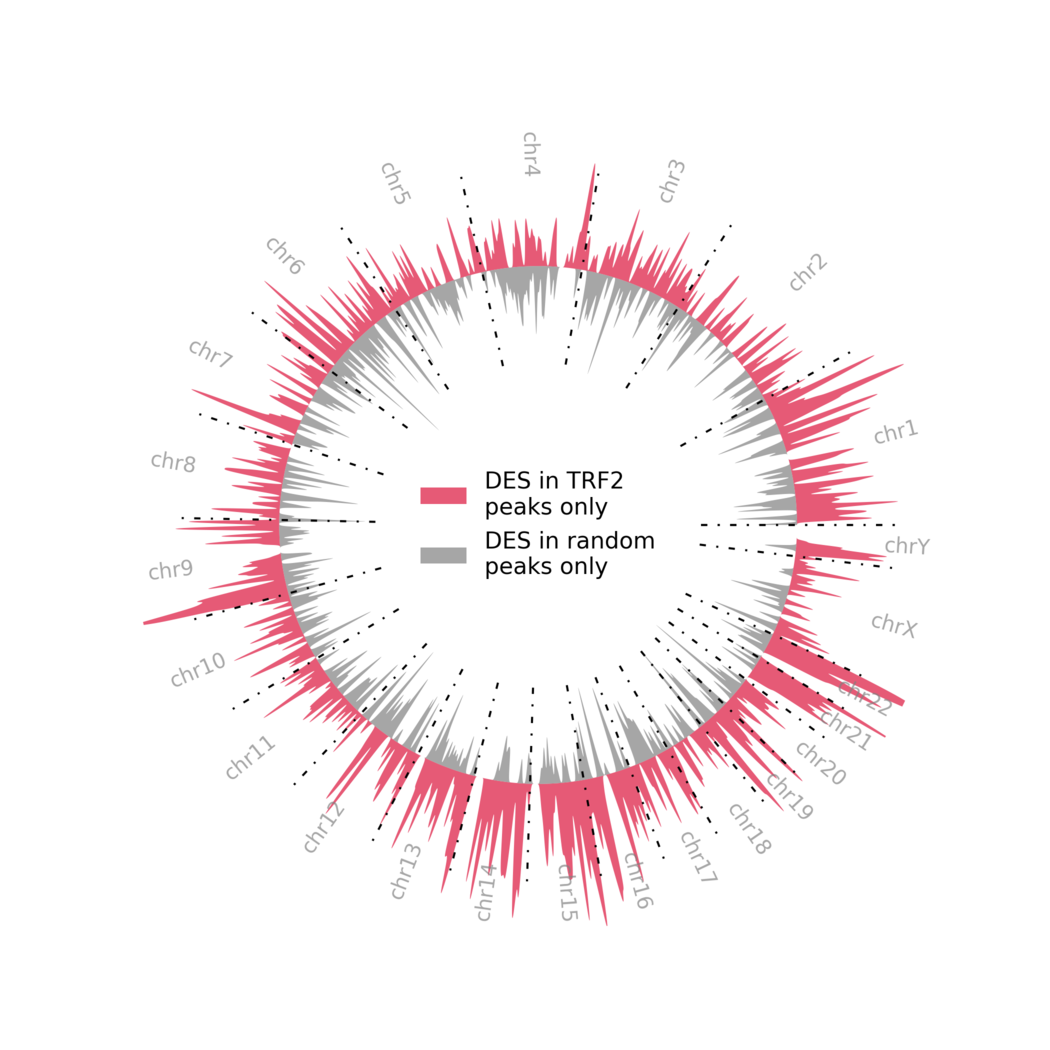
- DES differences between TRF2 and Random
python 2-d-Figure-2B-TRF2vsRandom.py
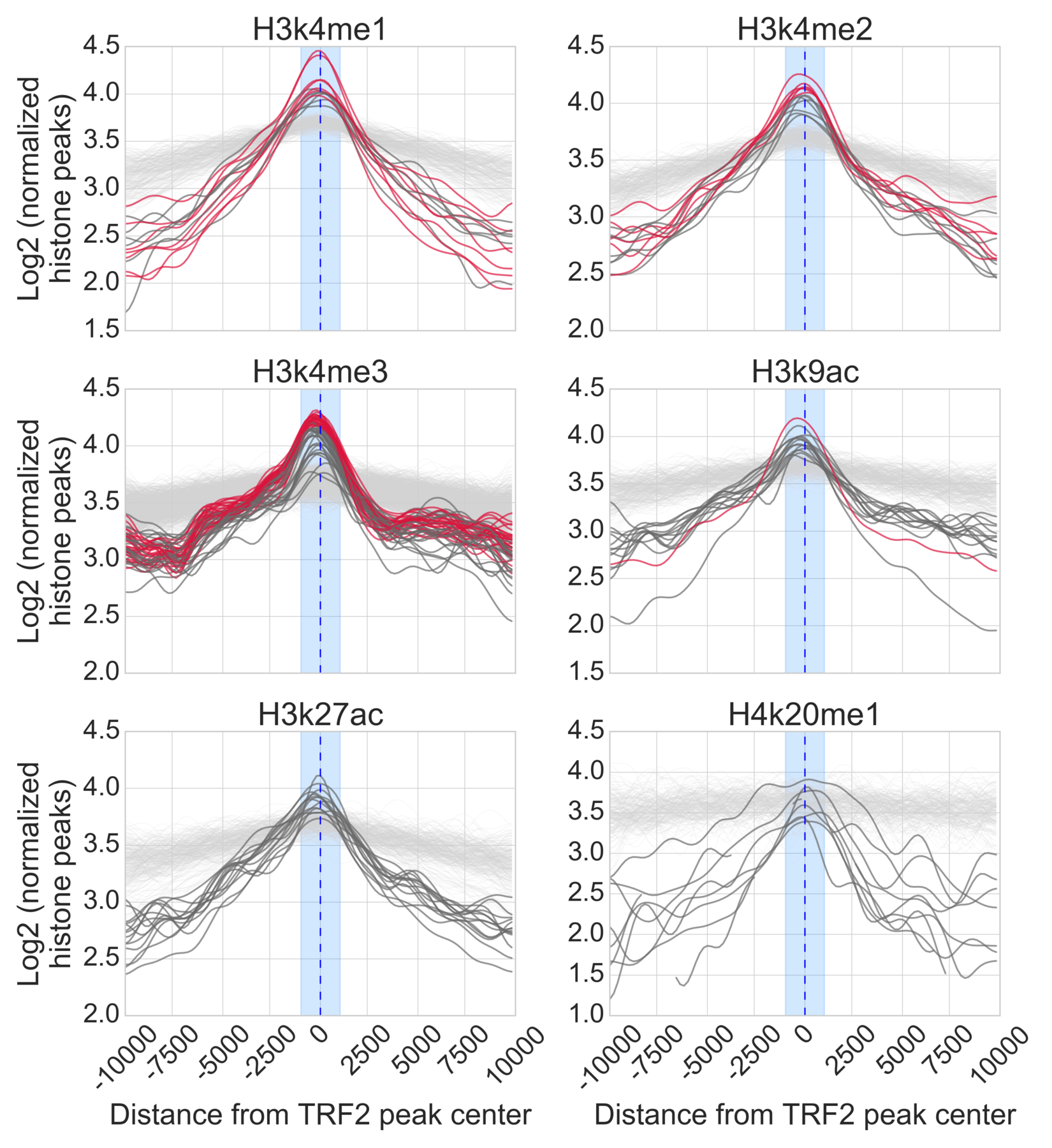
- Download Histone peaks from ENCODE
Rscript 3-a-download-ENCODE-Histone-marks.R
- Find closest mark
python 3-b-histones-closest-dist.py
- Format histone file names
Rscript 3-c-format-filename-histone-marks.R
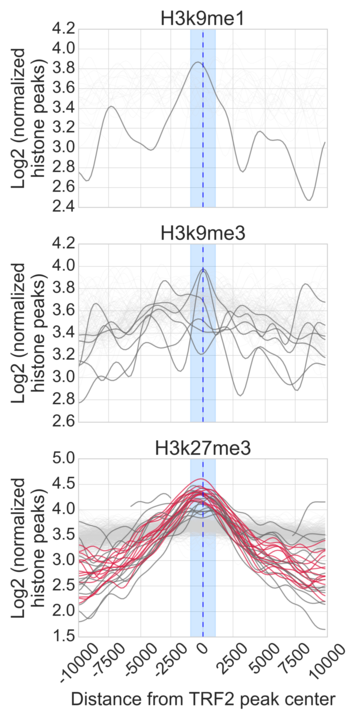
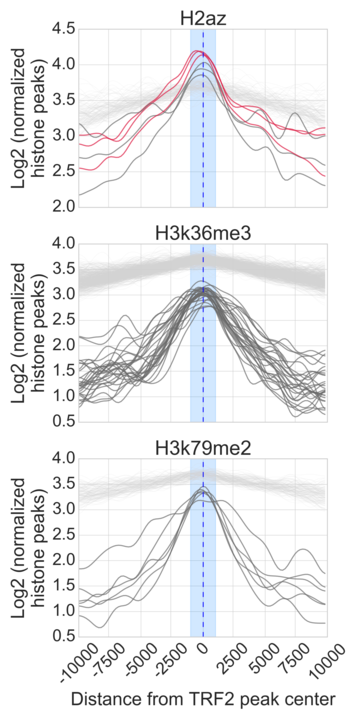
- Generate Figure 2A-C and supplementary figure 4A-D
python 3-d-generate-Fig-3A-C-and-Suppl-fig-4A-D.py