Pytorch implementation of Variational Network for Magnetic Resonance Image (MRI) Reconstruction
This repository contains a pytorch implementation of the variational network for MRI reconstruction that was published in these papers.
- Hammernik et al., Learning a variational network for reconstruction of accelerated MRI data, Magnetic Resonance in Medicine, 79(6), pp. 3055-3071, 2018.
- Knoll et al., Assessment of the generalization of learned image reconstruction and the potential for transfer learning, Magnetic Resonance in Medicine, 2018 (early view).
Please consider citing the original authors if you use this codes and data:
@article{doi:10.1002/mrm.26977,
author = {Hammernik Kerstin and Klatzer Teresa and Kobler Erich and Recht Michael P. and Sodickson Daniel K. and Pock Thomas and Knoll Florian},
title = {Learning a variational network for reconstruction of accelerated MRI data},
journal = {Magnetic Resonance in Medicine},
volume = {79},
number = {6},
pages = {3055-3071},
keywords = {variational network, deep learning, accelerated MRI, parallel imaging, compressed sensing, image reconstruction},
doi = {10.1002/mrm.26977},
url = {https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.26977},
eprint = {https://onlinelibrary.wiley.com/doi/pdf/10.1002/mrm.26977},
}
Sample reconstruction
Below are some sample reconstruction of 4 times under-sampled data from a model trained with coronal PD FS data.
 Sample coronal PD FS reconstruction
Sample coronal PD FS reconstruction
 Sample coronal PD reconstruction
Sample coronal PD reconstruction
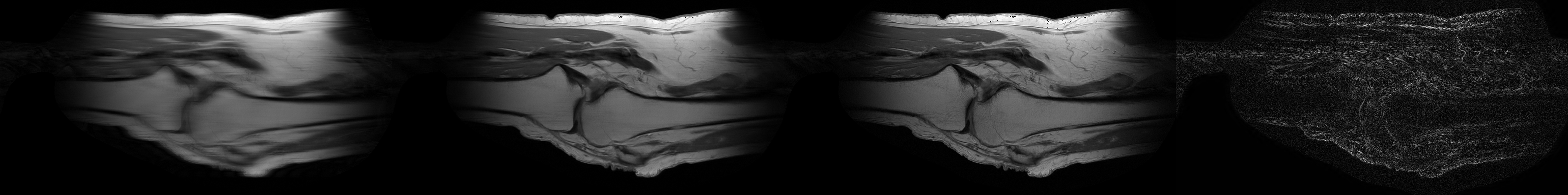 Sample Sagittal PD reconstruction
Sample Sagittal PD reconstruction
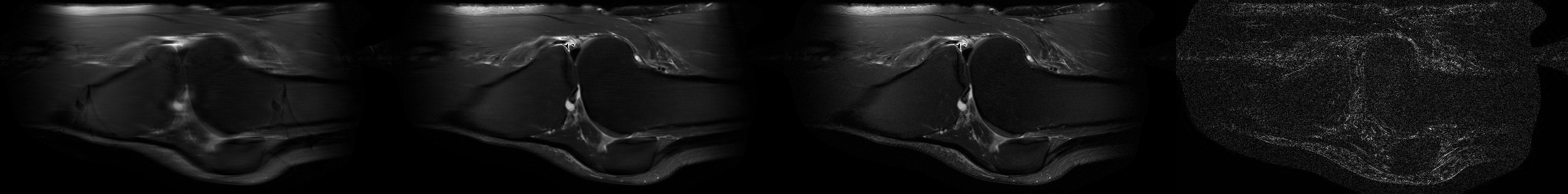 Sample Sagittal T2 reconstruction
Sample Sagittal T2 reconstruction
Requirements
The codes in this repo has been tested with Ubuntu 16.04, pytorch 1.6.0, and python 3.8.3 with anaconda. You can create an anaconda environment with the included env.yml to make sure the codes run without problems.
Data
The repo used the same data from the original authors, which can be accessed at GLOBUS.
The data loading and preprocessing step has been copied from the original repo and can be seen in the data_utils.py file. Some of the default parameters such as traing patients or testing slice range can be changed in the DEFAULT_OPTS variable.
Trainable parameters
The trainable parameters are the same as those of the original repo: the filters' kernels, the weights of the Gaussian RBF activation function, and the data term weight.
Training and testing
Training and testing can be done from the run_varnet.py script. For training, a variety of optimizers (SGD, Adam, RMSprop) can be used in addition to the IIPG optimizer (experimental) mentioned in the paper. Please check the script for a list of available parameters that can be modified.
Sample training command for coronal_pd_fs dataset:
python run_varnet.py --mode train --root_dir data/knee --name coronal_pd_fs --gpus 0 --epoch 50
By default, the training will save the model at the final epoch with the name varnet.h5 at exp/basic_varnet folder. Training progress is monitored with tensorboard at the lightning_logs/version_xx folder by default.
Sample testing command:
python run_varnet.py --mode eval --root_dir data/knee --name coronal_pd_fs --gpus 0
By default, the code will load the model varnet.h5 at exp/basic_varnet and run inference on the evaluation patients and slices defined at data_utils.py. The zero-filled input, network output, ground truth fully-sampled image, and error map is saved at the experiment directory.
A model trained on coronal PD FS data is provided in the trained_model folder.
Visualization
Visualization of the learnt kernel and the activation function can be run with the notebook visualization.ipynb. Path to a trained model needs to be supplied. The kernels are shown as (real,imaginary) pair. Activation and potential function for a specific cell and channel can be examined.
To-do
- Implement quantitative metrics (PSNR, SSIM)