Created: August 12, 2022 11:06 PM Project: Sepsis_Algorithm Reviewed: No
Developing sepsis prediction algorithm to detect acute deterioration.
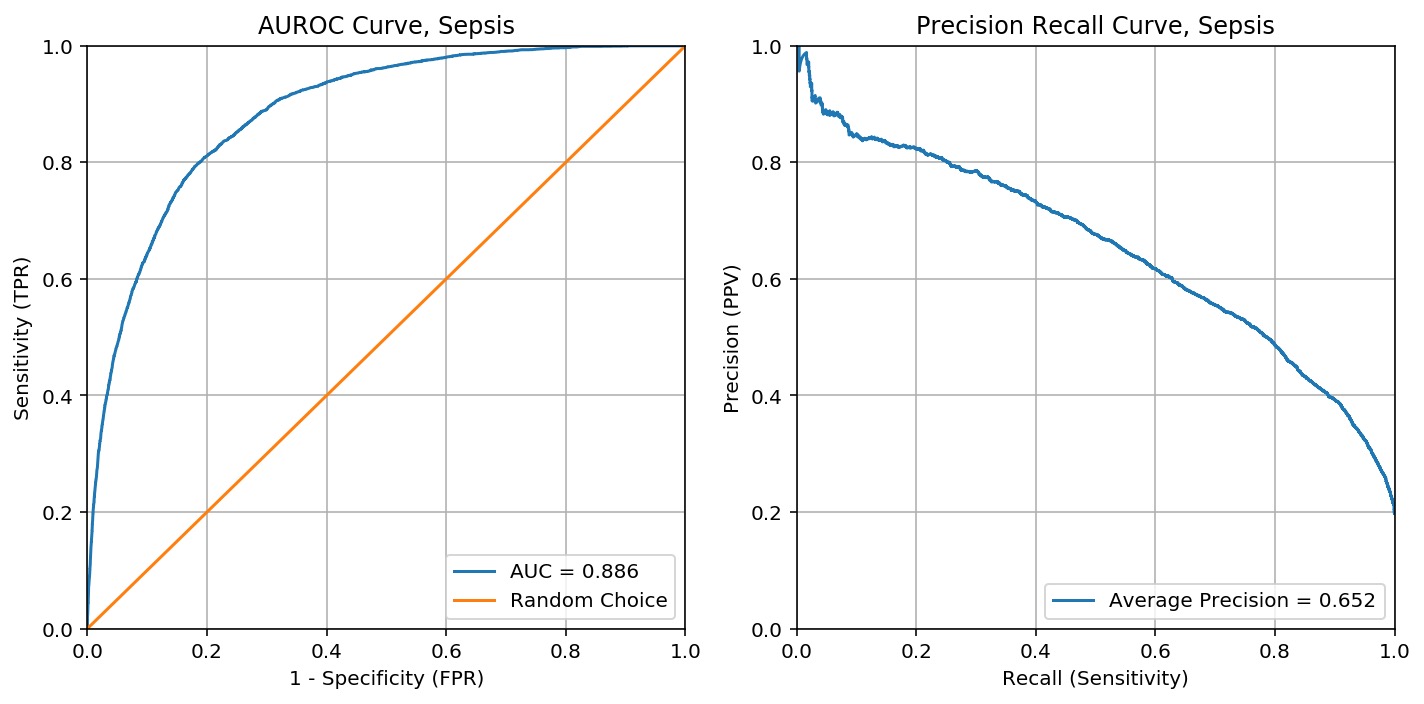
The Sepsis ML Model is an ensemble of five independently trained XGBoost. Each XGBoost model outputs a probability for developing sepsis every three hours using commonly collected vital signs and results from clinical lab testings, and the probabilities are averaged to obtain the final output.
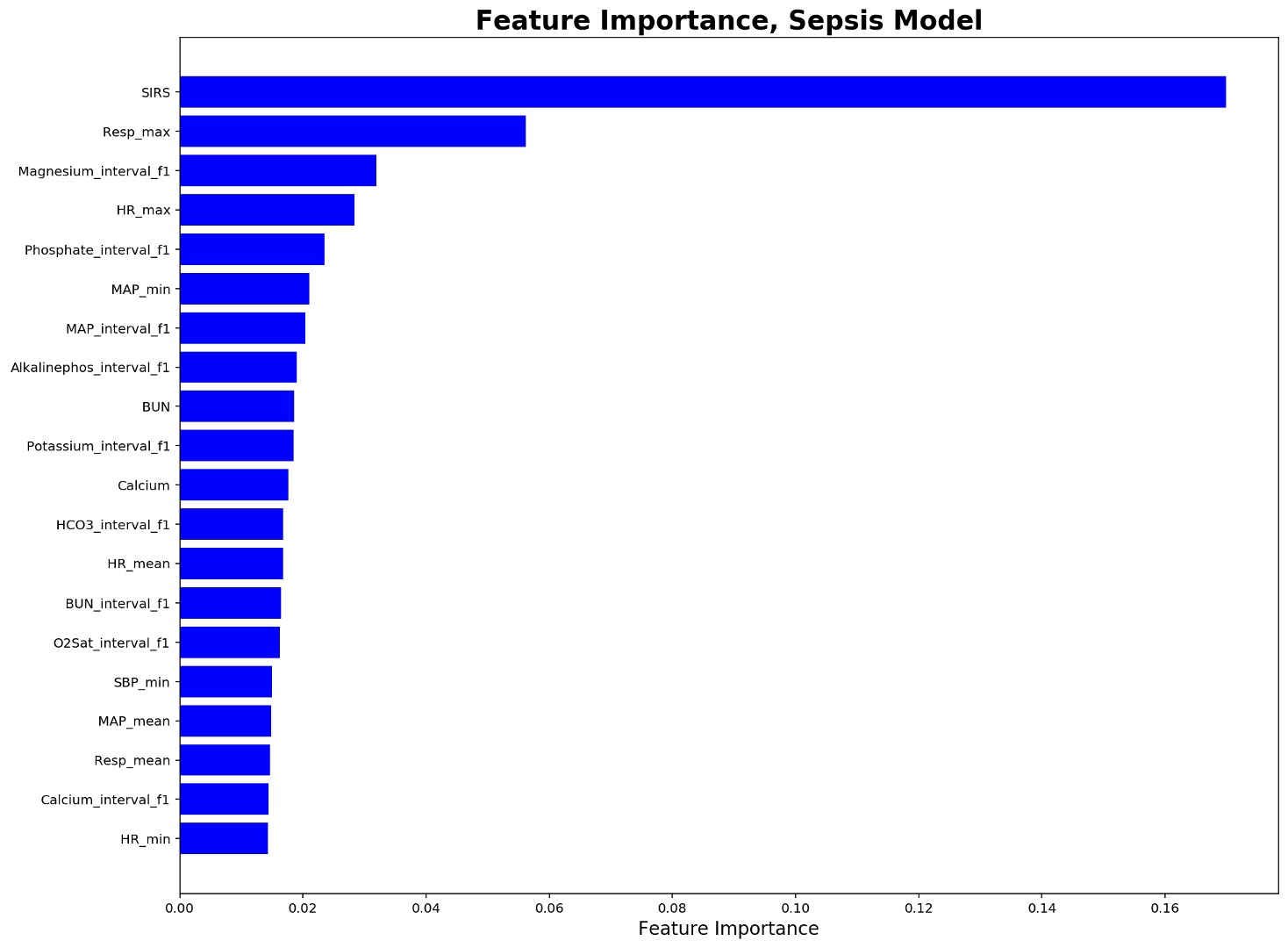
The input to the model has total of 174 features including all handcraft features inspired by https://github.com/Meicheng-SEU/EASP group’s work submitted to the Early Prediction of Sepsis from Clinical Data: The PhysioNet/Computing in Cardiology Challenge 2019.
- 36 Variables directly extracted from the EHR: followed dataset provided from the PhysioNet Challenge with few exceptions
- 9 Vitals: HR, O2Sat, Temp, SBP, MAP, DBP, Resp, EtCO2, GCS_total_score
- 25 Labs: AST, Alkalinephos, BUN, BaseExcess, Bilirubin_total, Calcium, Chloride, Creatinine, FiO, Glucose, HCO3, Hct, Hgb, Lactate, Magnesium, PTT, PaCO2, PaO2, Phosphate, Platelets, Potassium, SaO2, Sodium, WBC, pH
- 2 Demographics: age, gender (is_female)
- 30 Statistical features: 18-hr sliding window based statistical features
- _mean, _median, _min, _max, _std, _dstd
- derived for common vital signs (HR, O2Sat, Temp, Resp, SBP, MAP)
- 99 Missing value features & differential features
- 33 Measurement frequency sequence (_interval_f1): Records the number of variable measurements before the current time
- 33 Measurement time interval sequence (_interval_f2): Records the time interval from the last measurement between the current time.
- 33 Differential features (_diff): difference between the current value and the previous last known value
- derived for vitals (except gcs_total_score) & labs directly extracted from the EHR
- 9 Score quantified features
- HR_score, Temp_score, Resp_score, MAP_score, Creatinine_score, qsofa, Platelets_score, Bilirubin_score, SIRS
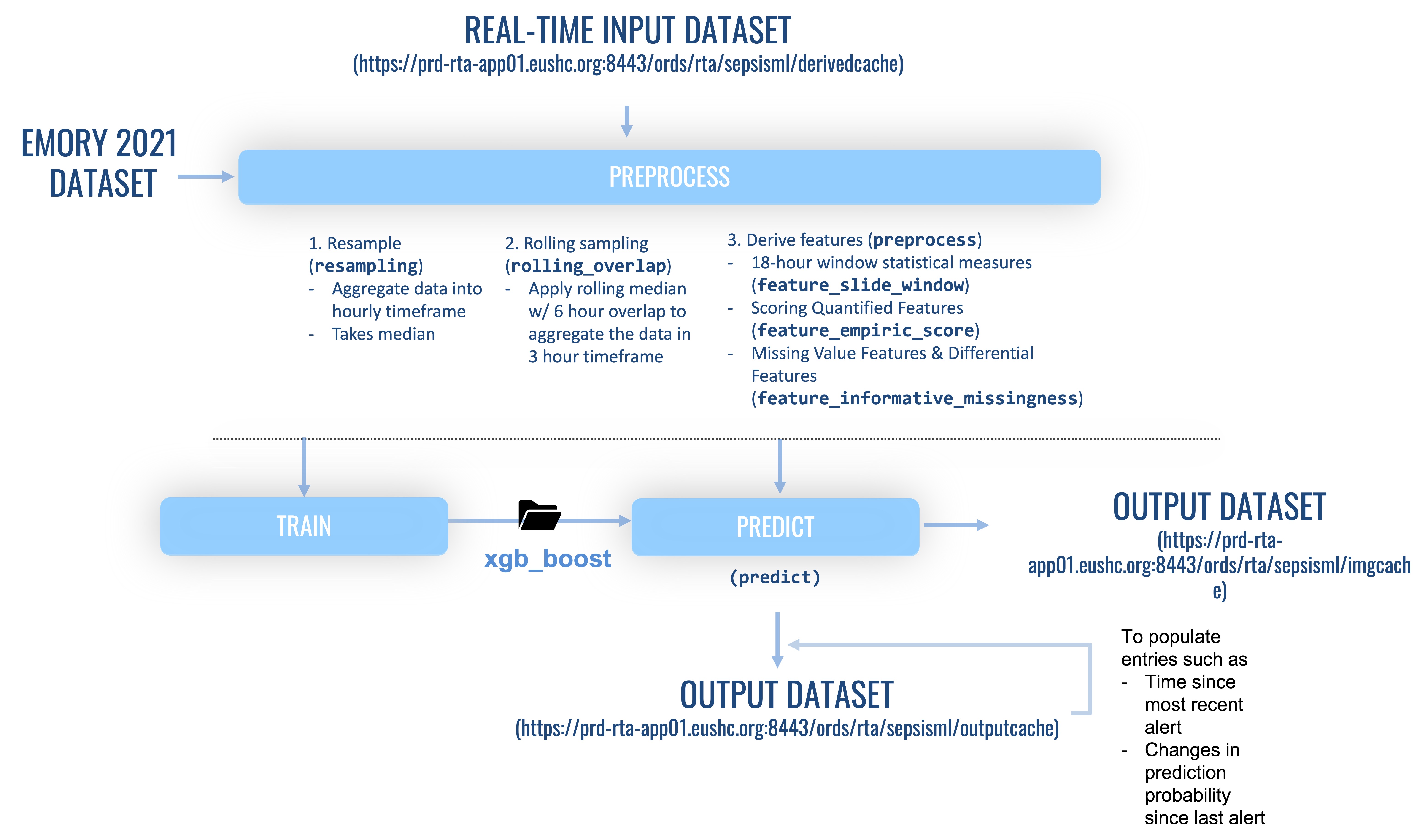
- Preprocess
- Resample sparse matrix to hourly longitudinal data
- Aggregate data into 3 hour bins with 6 hour sliding window
- Feature extraction/missing value imputation
- Train (retrospective data)
- RealTime Implementation (@ VM)
See the preprocess section from train_breakdown.ipynb or test_breakdown_ipynb for further explanation about the script
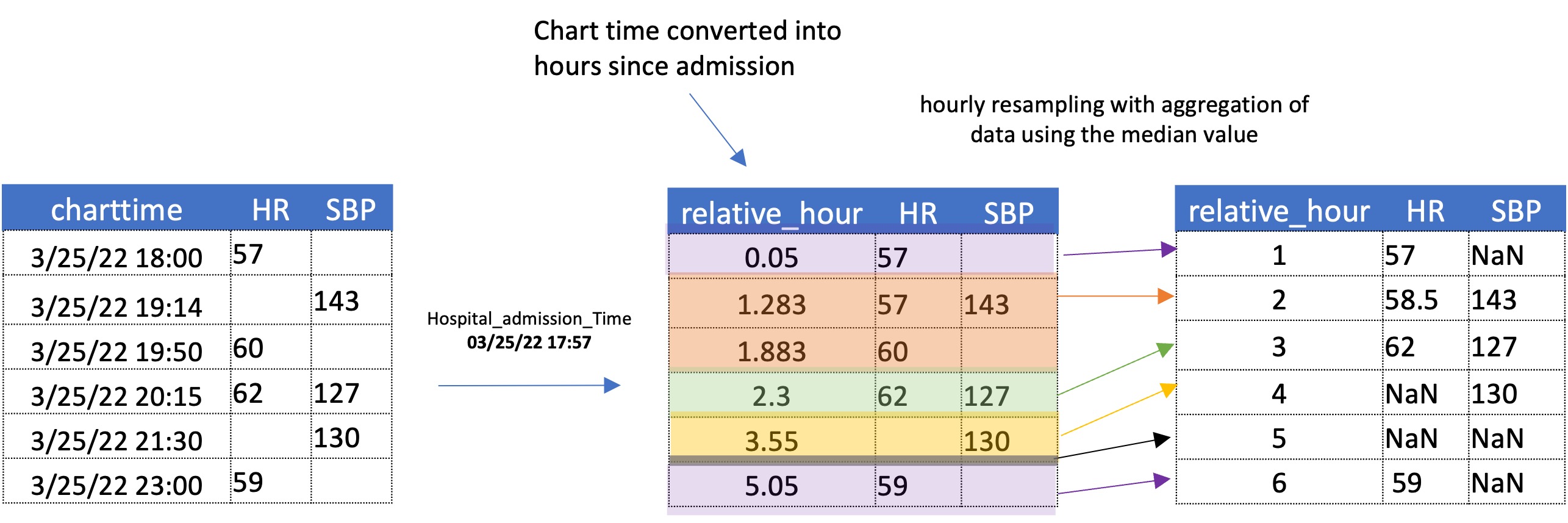
To obtain an hourly sampled dataset, data was resampled with respect to time of hospital admission while aggregating the data with its median value. The process is illustrated in the figure below with a simplified dataset.
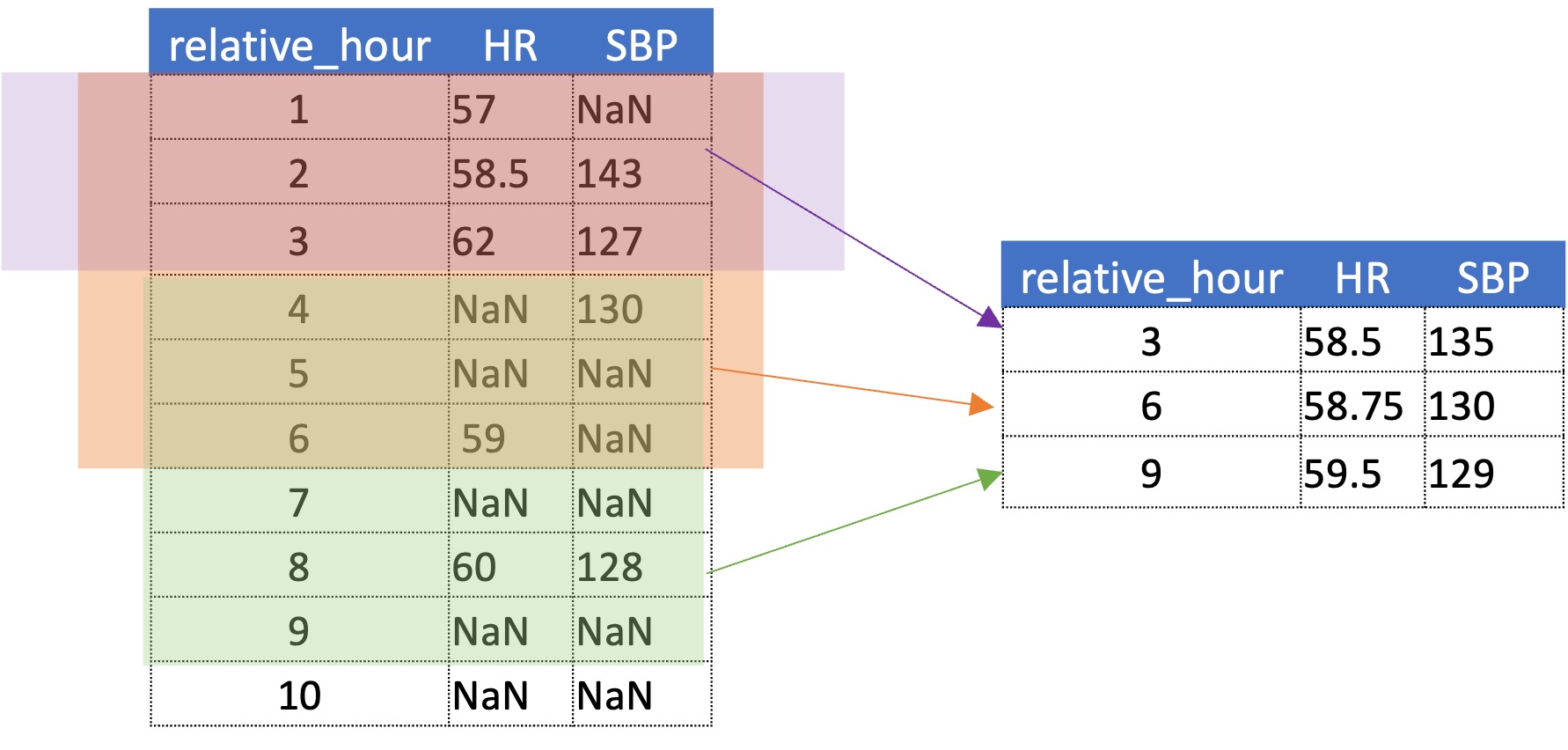
The hourly bin data is now aggregated into three hour bins with a 6-hour look-back (which creates a three hour overlap between previous entry)
Features including Statistical features, Missing value features & differential features, and Score quantified features are all extracted. While extracting the feature, the script also imputes the missing values with forward filling the last known value.
See the data_extraction_from_MODS.ipynb for further explanation about dataset extraction from MODS folder.
See the train_breakdown.ipynb for further explanation about training process.
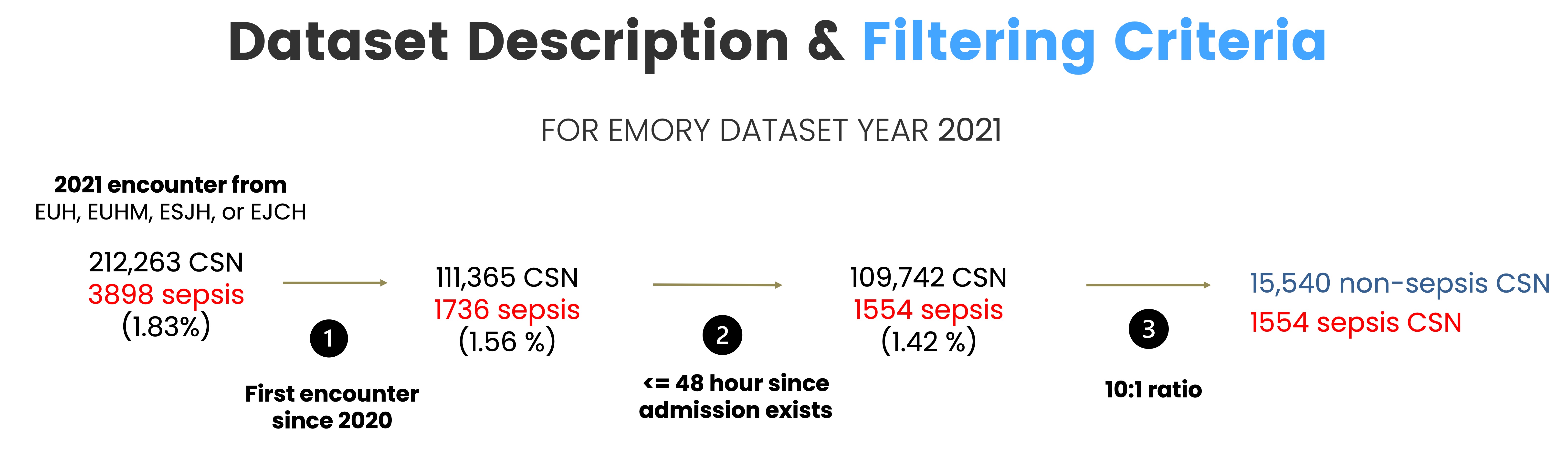
Model is train with retrospective data— Emory 2021 data. The following is the criteria used to define the cohort for training and validation.
5-fold training/validation was performed, creating five separate XGBoost models.
See the SepsisML_realtime_breakdown.ipynb for further explanation about the script
More data cleaning is required to match the dataset format from training, but similar preprocessing is performed on the data extracted from the API call. Here, the five pre-trained XGBoost models are called for testing.
- will be updated as soon as the most recent changes are implemented