This repository provides an introduction to basic techniques in genomic processing. Specifically, participants will learn an abbreviated and simplified overview of how raw reads can be converted to variants. The purpose of this introduction is to help individuals new to genomics understand the basics of file transformation; it is not to present every step in genomic data processing (additional steps are required).
- Understand major genomic filetype structure (.fastq, .sam, .vcf)
- Understand basic flow of a genomic bioinformatics pipeline
If you are here as a UTU student taking BIOL 3300, you should do the following:
-
Login to your Github account.
-
Fork this repository, by clicking the 'Fork' button on the upper right of the page.
After a few seconds, you should be looking at your copy of the repo in your own Github account.
-
Click the 'Clone or download' button, and copy the URL of the repo via the 'copy to clipboard' button. note: if you have an SSH key with your github account, make sure you select the
SSHtab -
In your terminal, navigate to where you want to keep this repo (you can always move it later, so just your home directory is fine). Then type:
$ git clone the-url-you-just-copiedand hit enter to clone the repository. Make sure you are cloning your fork of this repo.
-
Next,
cdinto the directory:$ cd the-name-of-directory-you-just-cloned -
At this point, you should be in your own local copy of the repository.
As you work on the exercise below, be sure to frequently
commityour work andpushchanges to the remote copy of the repo hosted on Github.
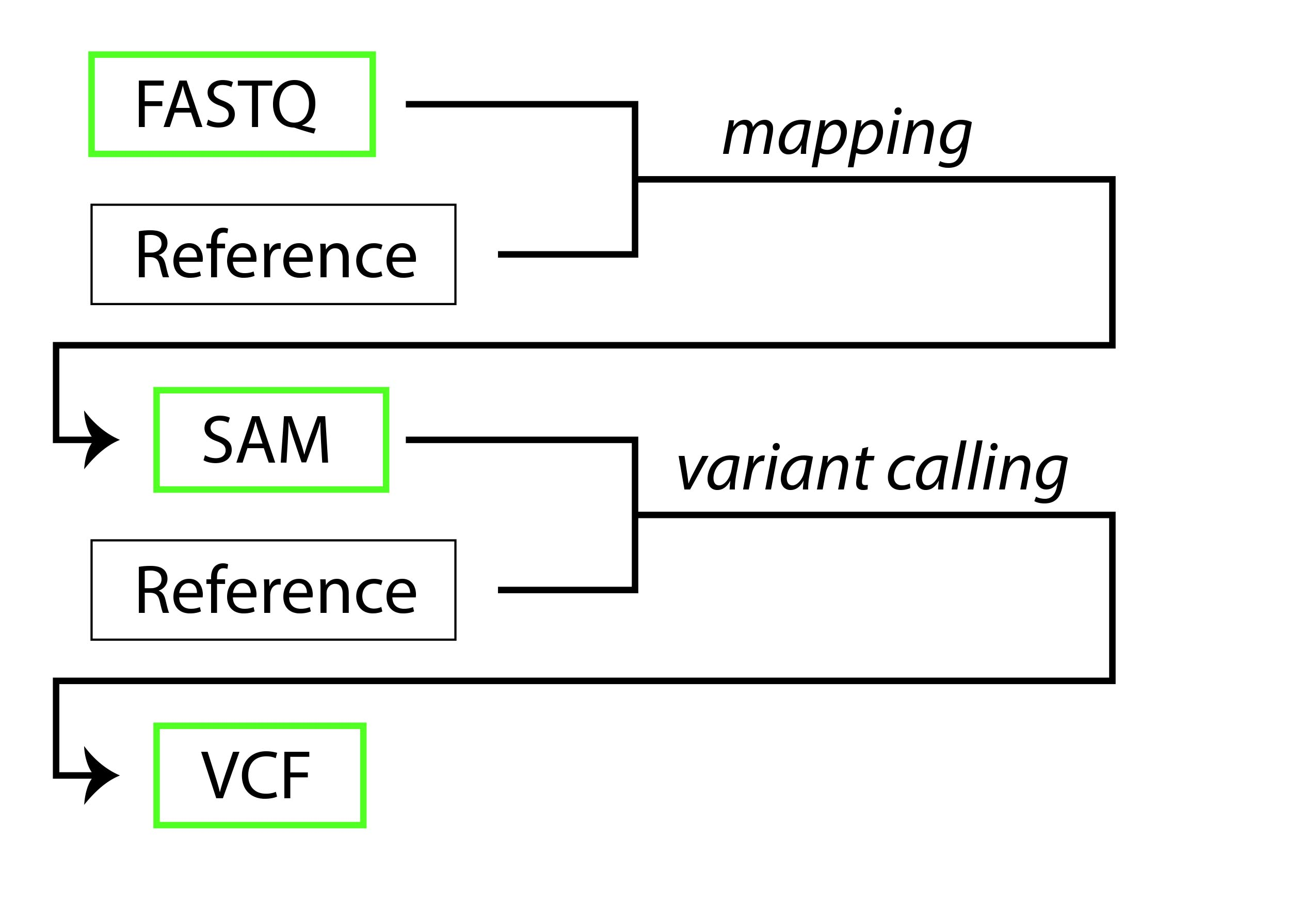
For this introduction, you will be introduced to three genomic filetypes:
The purpose of a bioinformatics pipeline is to transform biological sequence data to a format that can be interpreted. In many cases, researchers are interested in variable regions within the genomes (variants). To find these regions, raw sequencing reads must be aligned (mapped) to a reference, following which the variants can be determined. An abbreviated and simplified overview of file transformations via bioinformatics pipeline is as follows:
The "Reference" in the above diagram is an already-assembled FASTA sequence used to orient the data. The italicized words represent file transformations performed by bioinformatics software)
Perhaps the most common filetype to store genetic data, FASTA files simply contain sequence headers and the sequences. A fasta file can have a single header or multiple headers. The header line can contain various items pertaining to the sequence, such as the chromosome, species, and individual ID. A header line is demarcated by the '>' character, and the following line(s) contain the sequence information pertaining to this specific header. The sequence line should only contain information pertaining to the sequence described by the previous header. Some FASTA files have sequences that span multiple lines (these are called 'interleavened' files), and others have the entire sequence contained in a single line. Here is a sample of each:
Interleavened FASTA file:
>Chromosome_1, SARS-Cov2, Sample_1
ATTAAAGGTTTATACCTTCCCAGGTAACAAACCAACCAACTTTCGATCTCTTGTAGATCTGTTCTCTAAA
CGAACTTTAAAATCTGTGTGGCTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATTAATAAC
TAATTACTGTCGTTGACAGGACACGAGTAACTCGTCTATCTTCTGCAGGCTGCTTACGGTTTCGTCCGTG
TTGCAGCCGATCATCAGCACATCTAGGTTTCGTCCGGGTGTGACCGAAAGGTAAGATGGAGAGCCTTGTC
>Chromosome_1, SARS-Cov2, Sample_1
ATTAAAGGTTTATACCTTCCCAGGTAACAAACCAACCAACTTTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGGCTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATTAATAACTAATTACTGTCGTTGACAGGACACGAGTAACTCGTCTATCTTCTGCAGGCTGCTTACGGTTTCGTCCGTGTTGCAGCCGATCATCAGCACATCTAGGTTTCGTCCGGGTGTGACCGAAAGGTAAGATGGAGAGCCTTGTC
Both of these examples contain the same information: header line of '>Chromosome_1, SARS-Cov2, Sample_1' and a single nucleotide sequence (280 nucleotides long) starting with 'ATTAAA' and ending with 'CCTTGTC'
As stated above, the nucleotide sequences for reference genomes are stored in fasta files. As you can imagine, sometimes these files can be quite large. However, this is dependent on the size of the genome. The human genome contains over three billion base pairs, whereas viral genomes can contain just several thousand. Let's look at an example genome stored in a FASTA file: a SARS-CoV-2 genome. At the command line you can examine this file using vim covid19-refseq.fasta.
note: If you are new to using vim, you can exit without saving by typing ':q!' followed by enter.
Once you exit vim, you can count the number of sequences within this file by counting the number of headers:
grep '>' covid19-refseq.fasta | wc -l
The SARS-CoV-2 genome contains a single chromosome, thus the reference genome only has one header.
The filetype that is typically provided by a sequencing facility, FASTQ files are commonly referred to as "raw read" files. Similar to .fasta files, FASTQ files contain a sequence identifier and biological sequence data. Additionally, FASTQ files contain a sequencing quality score for each base pair position. Here is an example of one sequence and its associated information:
@SeqID
AAGCCAGCAAACCTTGTTTTACCTCACTGATATAGATTAGATATTTCAAGACAAATTTGTTGCCAATGTTAGATTATTAACATTATTTATTATAAAAATA
+
CCCFFFFFHHHHHJJJJJJJJJJJJJJJJIJJJJJJJJJJJJIJIJJJJJJJJJJJJJJJJJJJJJJIJJJJJJIJJJJHHHHHHHFFFFFFFEEEEEEC
The first line is the sequence identifier. The first character for the identifier line is '@'. Similar to the '>' character in .fasta files, the '@' character in FASTQ files denotes the sequence identity for the following sequence. Additionally, this line might contain a description of the sequence.
The second line contains the sequence itself (string of nucleotides). The sequence is followed by a '+' on the third line to indicate the end of the sequence string.
The fourth line contains a quality score for each position of the sequence. Each character represents a number based on ASCII coding (see this link for the relationship between symbols and quality score value). On this scale, 0 ('!') is the lowest value, and 40 ('I') is the highest value. Because each score corresponds to a site within the sequence itself, the number of score symbols must equal the number of positions in the sequence.
Let's look at an example FASTQ file. These files can be very large, but example.fastq is an abbreviated file that can be opened in your text editor. If on the command line, you can examine this file using vim example.fastq.
note: If you are new to using vim, you can exit without saving by typing ':q!' followed by enter.
You'll notice that the sequence identifier line is more complex than the example above. Sequencing companies use this line to provide unique characteristics of each sequence. For example, Illumina paired-end sequencing (the platform and method used to obtain this sequencing data) uses a specific format for the sequence ID and description.
With this info, you can parse out the information from the first sequence id in example.fastq as follows:
| Order | description | value |
|---|---|---|
| 1 | instrument | D3NH4HQ1 |
| 2 | run number | 149 |
| 3 | flowcell ID | C1H5KACXX |
| 4 | lane | 3 |
| 5 | tile | 1101 |
| 6 | x-pos | 2106 |
| 7 | y-pos | 2242 |
| - | space | - |
| 8 | read | 2 |
| 9 | is filtered | N |
| 10 | control number | 0 |
| 11 | index number | GCTCGGTA |
For the purposes of this introduction, you don't need to worry about all of these elements– just that this line is the unique identifier for the sequence with additional sequencing details.
Sequence alignment map (SAM) files are text-based genomic files with biological sequence data aligned to a reference sequence. SAM files contain a header section and an alignment section. They contain the same information as the FASTQ file, with additional information on mapping details. As you probably gathered, that makes these files larger than the FASTQ files. To increase computational efficiency, SAM files can be converted into a binary alignment Map (BAM) file. BAM files are much smaller than SAM files, and this conversion is commonly done in genomic processing. Here is an example of header and alignment lines within a SAM file:
@SQ SN:NC_045541.1 LN:186725308
@PG ID:bwa PN:bwa CL:bwa mem -t 4 -M RefGenome.fasta Read1.fastq Read2.fastq
SeqID 99 ref_name 72165682 60 100M = 72165982 399 TACTTATGTTCTTCTTCATTCAGGATCATATGTGAAACTTCAGAAAAGCTAATATGTGAAACTTCAGAAGACAAATATGGTGAGAACAACAGTGAAAGAG CCCFFFFFHHHHHJIJJJJJJJIJIJJJJJIJJJJJJJJJJIIJGIJJJJJJJIJJBGIJJJIJIIJJIJJJIIICFIHEA@EGHHHHHFFFEFEEEEDE
The header section precedes the alignment section, and each heading begins with the '@' symbol. Each heading contains tab-delimited sections. The first column indicates the record type. The following columns contain tags and values (in the format TAG:VALUE). While there are different tag types, two you will see often are @SQ (reference sequences) and @PG (programs used for creating .sam). The values of these tags contain information about the sequence. @SQ requires the reference sequence name (SN) and length (LN) tags, and the @PG tag requires the program identity but may also include the program name (PN), version (VN), and command line implementation (CL).
The alignment section requires 11 tab-separated fields, and additional fields are optional. Each line within this section represents the alignment of a segment to the reference. The 11 required sections include information on the query template (read that mapped), the reference sequence name (SN), the position on reference where the query template mapped, the mapping quality, the sequence itself, and the quality score for each position in the base pair. Simplified descriptions of each required field are within the table in the looking at a .SAM file section.
Let's look at an example SAM file. These files can be very large, but example.sam is an abbreviated file that can be opened in your text editor. If on the command line, you can examine this file using vim example.sam.
note: If you are new to using vim, you can remove text wrap by typing ':set nowrap' followed by enter. You can see line numbers by typing ':set number' followed by enter. You can exit vim without saving by typing ':q!' followed by enter.
You'll see that there are many @SQ header lines (one for each of the reference sequences). Each of these has a name and length. At line 366 you'll see the @PG header line for the program details. The remaining lines of the file contain alignment information. From what we learned above, we can parse the first alignment line (line 367) as follows:
| Col | Field | Type | Description | Value |
|---|---|---|---|---|
| 1 | QNAME | string | query template name | D3NH...:4262:2214 |
| 2 | FLAG | int | bitwise flag | 99 |
| 3 | RNAME | string | ref sequence name | NC_045541.1 |
| 4 | POS | int | 1-based leftmost mapping position | 72165682 |
| 5 | MAPQ | ing | mapping quality | 60 |
| 6 | CIGAR | string | CIGAR string | 100M |
| 7 | RNEXT | string | ref name of the mate/next read | = |
| 8 | PNEXT | int | position of the mate/next read | 72165982 |
| 9 | TLEN | int | template length | 399 |
| 10 | SEQ | string | segment sequence | TACTTATGTTCT... |
| 11 | QUAL | string | ASCII score of base quality | @DCC?CCEC>CE... |
Variant call format (VCF) files are text-based genomic files with information on sequence variation. More specifically, it includes sites where multiple characters are present in the samples examined. A VCF file contains a header section and a variant data section. Basic VCF files do not contain information on every position from the FASTQ or reference file, rather they include information on the genomic positions with sequence variation. As you probably gathered, that makes these files smaller than the FASTQ and SAM files (and the less variation, the smaller the file). Here is an abbreviated example of header and alignment lines within a VCF file:
##fileformat=VCFv4.2
##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">
##FORMAT=<ID=AD,Number=R,Type=Integer,Description="Allelic depths for the ref and alt alleles in the order listed">
##FORMAT=<ID=DP,Number=1,Type=Integer,Description="Approximate read depth (reads with MQ=255 or with bad mates are filtered)">
##FORMAT=<ID=GQ,Number=1,Type=Integer,Description="Genotype Quality">
##INFO=<ID=AC,Number=A,Type=Integer,Description="Allele count in genotypes, for each ALT allele, in the same order as listed">
##INFO=<ID=AF,Number=A,Type=Float,Description="Allele Frequency, for each ALT allele, in the same order as listed">
##INFO=<ID=DP,Number=1,Type=Integer,Description="Approximate read depth; some reads may have been filtered">
##INFO=<ID=FS,Number=1,Type=Float,Description="Phred-scaled p-value using Fisher's exact test to detect strand bias">
##contig=<ID=NC_045541.1,length=186725308,assembly=reference.fasta>
##reference=file://reference.fasta
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT Sample01 Sample02 Sample03
NC_045541.1 1206 . A G 138.21 . AC=2;AF=0.25;DP=6;FS=0.000 GT:AD:DP:GQ 0/0:6,0:6:42 ./.:0,0:0:0 1/0:5,7:12:71
The header section precedes the variant data section, and each heading begins with '##' symbols (notice there are two hash marks). Information about the variant dataset, the reference sequence, and the program used to generate the VCF are contained within the header.
The FORMAT header lines define tags whose properties pertain to the variant site as a whole, whereas the INFO header lines describe tags whose properties pertain to the genotype for each individual in the dataset. The abbreviations in FORMAT and INFO header lines correspond with those in the data section. The abbreviated VCF file example above defines four FORMAT tags (GT, AD, DP, and GQ) and four INFO (AC, AF, DP, and FS) tags. The contig and reference sections contain information about the reference used for variant calling.
The variant section consists of a row for every variant. The columns provide information about (1) site-level properties and annotations and (2) sample-leve annotations. The first section of columns (site-level properties and annotations) correspond to the variant site as a whole. This section consists of 8 columns, all of which are required in the vcf file (CHROM, POS, ID, REF, ALT, QUAL, FILTER, INFO). These required fields include information about the location, the reference allele, the alternate allele(s), and the quality of the SNP. While these fields are required for each variant, they can be empty (a signified by .). Descriptions for each of these fields are shown in the table below (values are from the variant of the abbreviated vcf file above).
| Col | Field | Description | Value |
|---|---|---|---|
| 1 | CHROM |
Contig name | NC_045541.1 |
| 2 | POS |
Position of variant within contig | 1206 |
| 3 | ID |
Optional identifier for variant | . |
| 4 | REF |
Reference allele (sequence character(s) at POS in reference) | A |
| 5 | ALT |
Alternate allele (sequence character in at POS in at least one sample) | G |
| 6 | QUAL |
Phred-scaled probability that variant exists at this site given data* | 138.21 |
| 7 | FILTER |
PASS means the variant has passed filtering, . means no filtering has occurred |
. |
| 8 | INFO |
Site-level annotations (properties of variant site as a whole) | AC=2;AF=0.25;DP=6;FS=0.000 |
The INFO values correspond to the flags defined in the header, where descriptions are provided. In the abbreviated vcf file, we see that this variant as an allele count (AC) of 2 (there are two alleles at this site), a minor allele frequency (AF) of 0.4 (the alternate allele's frequency in the dataset), a depth of coverate (DP) of 6 (the average depth per-individual at this site is 6 reads), and a p-value (FS) of 0.000. There are typically more property fields than this in a vcf, but this hopefully gives you a sense of how to read these sections.
The subsequent columns pertain to sample-level annotations. These fields consist of the formatting for the sample-specific property (FORMAT) followed by a column for each sample. In the abbreviated vcf file above, there are three samples (Sample01, Sample02, and Sample03). The values within the sample columns correspond to the ordered flags shown in the FORMAT column. These properties are shown in table format below; the Col values are a continuation from the table above.
| Col | Field | Description | Value |
|---|---|---|---|
| 9 | FORMAT |
Format for sample-specific annotations | GT:AD:DP:GQ |
| 10 | Sample01 |
The annotation values for Sample 01 | 0/0:6,0:6:42 |
| 11 | Sample02 |
The annotation values for Sample 02 | ./.:0,0:0:0 |
| 12 | Sample03 |
The annotation values for Sample 03 | 1/0:5,7:12:71 |
The value column can be somewhat challenging to understand, so we'll break it down:
| Flag | Description | Sample01 |
Sample02 |
Sample03 |
|---|---|---|---|---|
| GT | Genotype* | 0/0 | ./. | 1/0 |
| AD | Allele depth** | 6,0 | 0,0 | 5,7 |
| DP | Total depth at variant site | 6 | 0 | 12 |
| GQ | Genotype quality*** | 42 | 0 | 71 |
** 0/0 = homozygous for ref allele; 1/1 = homozygous for alt allele; 1/0 = heterozygous; ./. no data
Now check out the example.vcf file. These files can be very large, but example.vcf is an abbreviated file that can be opened in your text editor. If on the command line, you can examine this file using vim example.vcf.
note: If you are new to using vim, you can remove text wrap by typing ':set nowrap' followed by enter. You can see line numbers by typing ':set number' followed by enter. You can exit vim without saving by typing ':q!' followed by enter.
Once you have exited the text editor, you can count the number of variants in the VCF from the command line using the following command:
grep -v '#' example.vcf | wc -l
There are 1597 variants contained within this VCF file.
The basic workflow and data for this exercise come from Farkas et al., 2021 and the associated github repository.
Download and analyze a small sample of genomic data using published scripts to see an applied process of genomic data processing.
To complete this exercise, complete the following steps and answer the questions contained within the worksheet.md file.
note: if you haven't cloned this repository yet, make sure you have it cloned (see Getting set up section)
- Make sure mini/anaconda and python versions = 2.7 and >=3.0 are installed.
- Make sure you are in repository directory and activate conda environment
conda config --add channels conda-forge # add conda-forge channel (if you haven't already done so)
conda config --add channels bioconda # add bioconda channel (if you haven't already done so)
conda env update --file environment.yml # install required programs
conda activate genomics-pipeline-intro # activate genomics-pipeline-intro enviroment
- Install the latest version of sra toolkit. See instructions here:
https://github.com/ncbi/sra-tools/wiki/02.-Installing-SRA-Toolkit
For this exercise, you will run a bash script containing an abbreviated version of the genomics processing pipeline from Farkas et al., 2021.
You can see the parameters required for the script by looking at the help menu:
bash bash_scripts/genomics-pipeline-intro.sh -h
Now that you have examined the script, run it.
bash bash_scripts/genomics-pipeline-intro.sh -l July_28_2020_NorAm.txt -g covid19-refseq.fasta -a 0.4999 -t 4
You should see messages printing to stdout as the script runs. The first of these messages will look like this:
Downloading SRA files from the given list of accessions
2022-03-19T16:00:22 prefetch.2.11.2: Current preference is set to retrieve SRA Lite files with simplified base quality scores.
2022-03-19T16:00:23 prefetch.2.11.2: 1) Downloading 'SRR11851929'...
2022-03-19T16:00:23 prefetch.2.11.2: SRA Normalized Format file is being retrieved, if this is different from your preference, it may be due to current file availability.
2022-03-19T16:00:23 prefetch.2.11.2: Downloading via HTTPS...
2022-03-19T16:00:26 prefetch.2.11.2: HTTPS download succeed
2022-03-19T16:00:26 prefetch.2.11.2: 'SRR11851929' is valid
2022-03-19T16:00:26 prefetch.2.11.2: 1) 'SRR11851929' was downloaded successfully
2022-03-19T16:00:26 prefetch.2.11.2: 'SRR11851929' has 0 unresolved dependencies
note: Running into error messages is part of the coding experience. If you run into an error message, don't get too discouraged! First, read the error message. Is it an easy fix? If you aren't sure, copy and paste the error message into an online search engine to see if anyone else has experienced the error and discovered a solution. If you continue having trouble, reach out to your instructor over email to ask for help.
The script will take a few minutes to run. Once the finished, check that everything ran to completion. Your directory should now contain 8 .fastq.gz files, 8 .bam files, 8 VCF files beginning with SRR, and 1 merged.vcf file. You can verify this information with the following commands:
ls *.fastq.gz | wc -l # Number of .fastq.gz files
ls -l *.fastq.gz # Make sure the .fastq.gz files aren't empty
# note: The fifth column of this output is the file size in bytes
ls *.bam | wc -l # Number of .bam files
ls -l *.bam # Make sure the .bam files aren't empty
ls SRR*.vcf | wc -l # Number of SRR*.vcf files
ls -l SRR*.vcf # Make sure the *.vcf files aren't empty
ls -l merged.vcf # Make sure the merged.vcf file isn't empty
grep -v "#" merged.vcf | wc -l # Count the number of SNPs within merged.vcf
If the above check worked, congratulations! You successfully ran a published bioinformatics pipeline! Once again, this is not a complete bioinformatics pipeline. The sampling was significantly reduced to allow for shorter computation time, and downstream variant processing is required.
Once you have completed the worksheet, add, commit, and push the worksheet and the logfile to your forked repository.
add worksheet.md logfile
git commit -m "ran script and answered worksheet questions"
git push