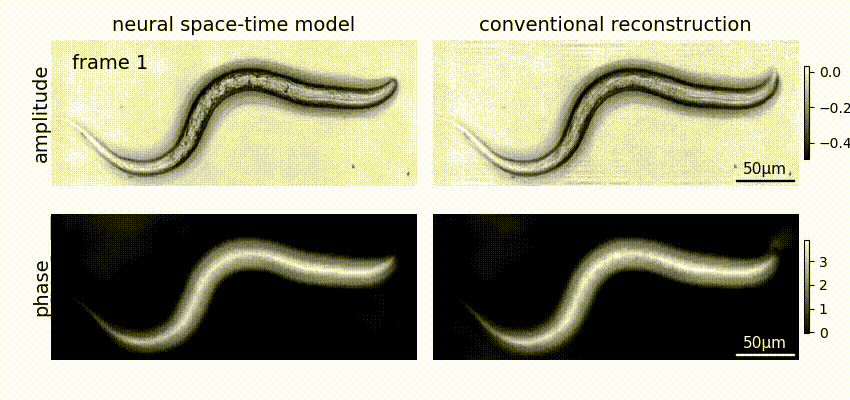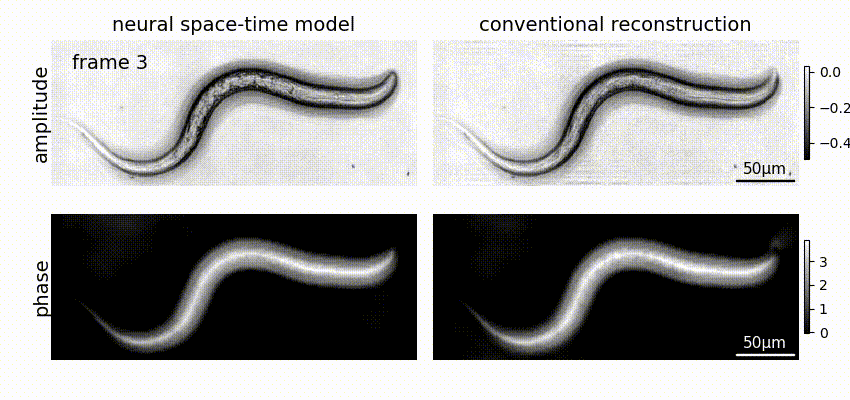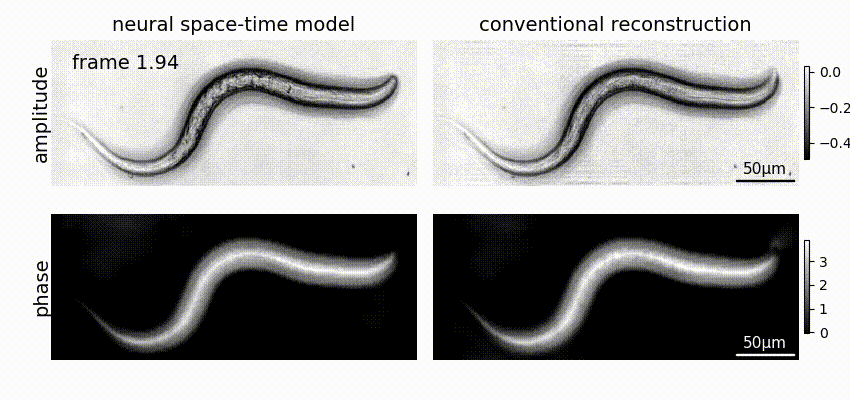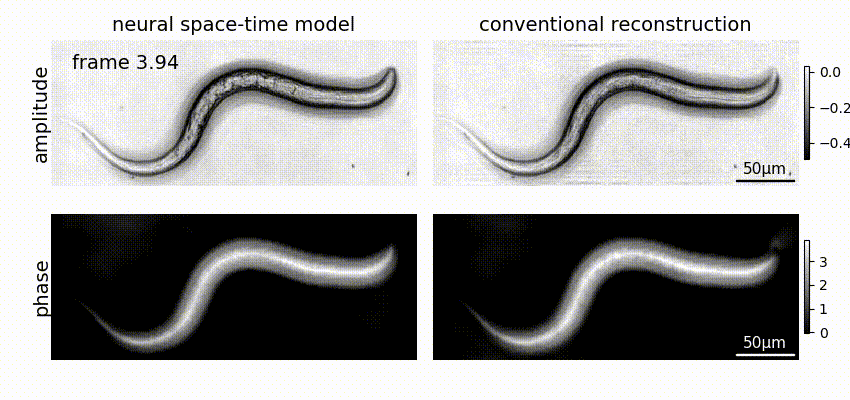
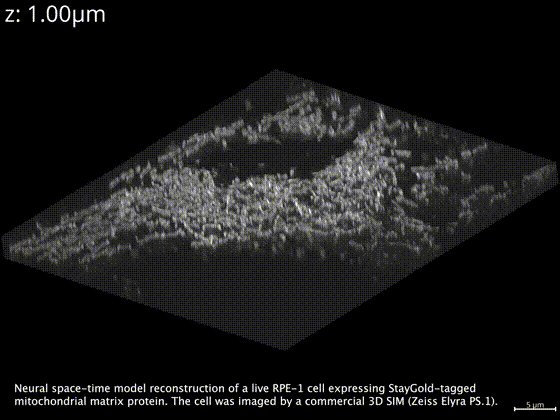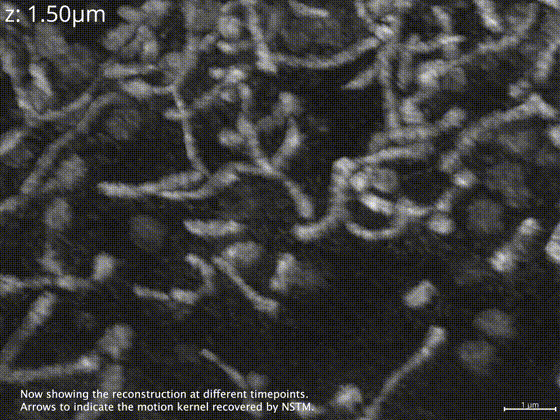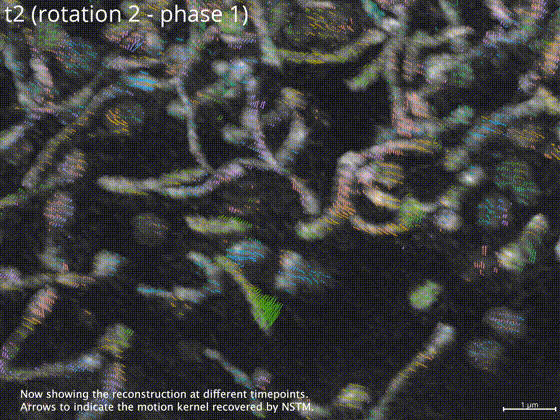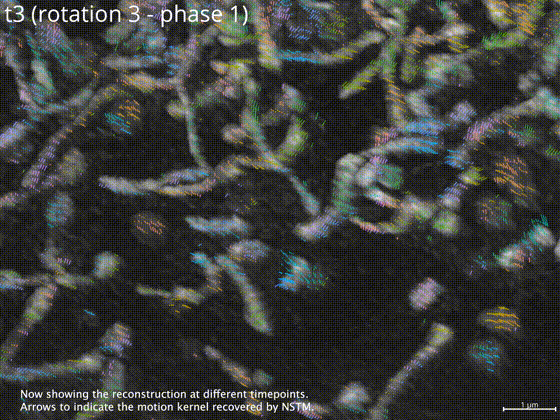
Neural space-time model (NSTM) is a computational image reconstruction framework that can jointly estimate the scene and its motion dynamics by modeling its spatiotemporal relationship, without data priors or pre-training. It is especially useful for multi-shot imaging systems which sequentially capture multiple measurements and are susceptible to motion artifacts if the scene is dynamic. Neural space-time model exploits the temporal redundancy of dynamic scenes. This concept, widely used in video compression, assumes that a dynamic scene evolves smoothly over adjacent timepoints. By replacing the reconstruction matrix, neural space-time model can remove motion-induced artifacts and resolve sample dynamics, from the same set of raw measurements used for the conventional reconstruction.
The usage of NSTM is demonstrated through three example imaging systems: differential phase contrast microscopy, 3D structured illumination microscopy, and rolling-shutter DiffuserCam. And there's a guide to incorporate NSTM into your own imaging system!
See the full documentation here and the original paper here.
Installation instructions are available in the documentation.
Locally run the step-by-step example in this Jupyter notebook or run on Google Colab
jupyter lab --notebook-dir=./nstm/examples
Option 1: Step-by-step example on Jupyter notebook with dense microbead data. You may also run this on Google Colab
Option 2: Run the python script for reconstruction
0. Download additional data from Google Drive and place .npz files in examples folder.
-
Start running endoplasmic reticulum (ER)-labeled cell reconstruction in commandline. Replace
er_cellwithmito_cellfor mitochondria-labeled cell data.python nstm/sim3d_main.py --config er_cellThe
mito_cellreconstruction takes ~40 minutes (slightly faster forer_cell) on a single NVIDIA A6000 GPU (48GB).er_cellis also runnable on a single NVIDIA RTX 3090 GPU (24GB) whenbatch_sizeis set to 1 in the .yaml file.mito_cellrequires close to 40GB GPU memory to run, as it has more image planes. -
The reconstruction results will be saved in
examples/checkpoint/folder. The 3D reconstruction volume with three timepoints (each corresponding to an illumination orientation) will be saved asrecon_filtered.tif, and can be viewed using Fiji. The recovered motion map will be saved asmotion_dense_t.npy. -
Additional reconstruction parameters are stored in
examples/configs/er_cell.yamlandexamples/configs/mito_cell.yaml. To print the full parameter descriptions, run:python nstm/sim3d_main.py --helpfull
@article{cao2024neural,
title={Neural space--time model for dynamic multi-shot imaging},
author={Cao, Ruiming and Divekar, Nikita S and Nu{\~n}ez, James K and Upadhyayula, Srigokul and Waller, Laura},
journal={Nature Methods},
pages={1--6},
year={2024},
publisher={Nature Publishing Group US New York}
}