[Update]--2023/06/09--for weights trained with GTEx v8, please visit https://zhaocenter.org/UTMOST.
[Update]--2018/04/24--pre-calculated covariance matrices for single-tissue and joint tests are downloadable now; updated pipeline for single-tissue/joint tests using 44 GTEx tissues + STARNET liver + BLUEPRINT 3 cell types (eQTL/sQTL).
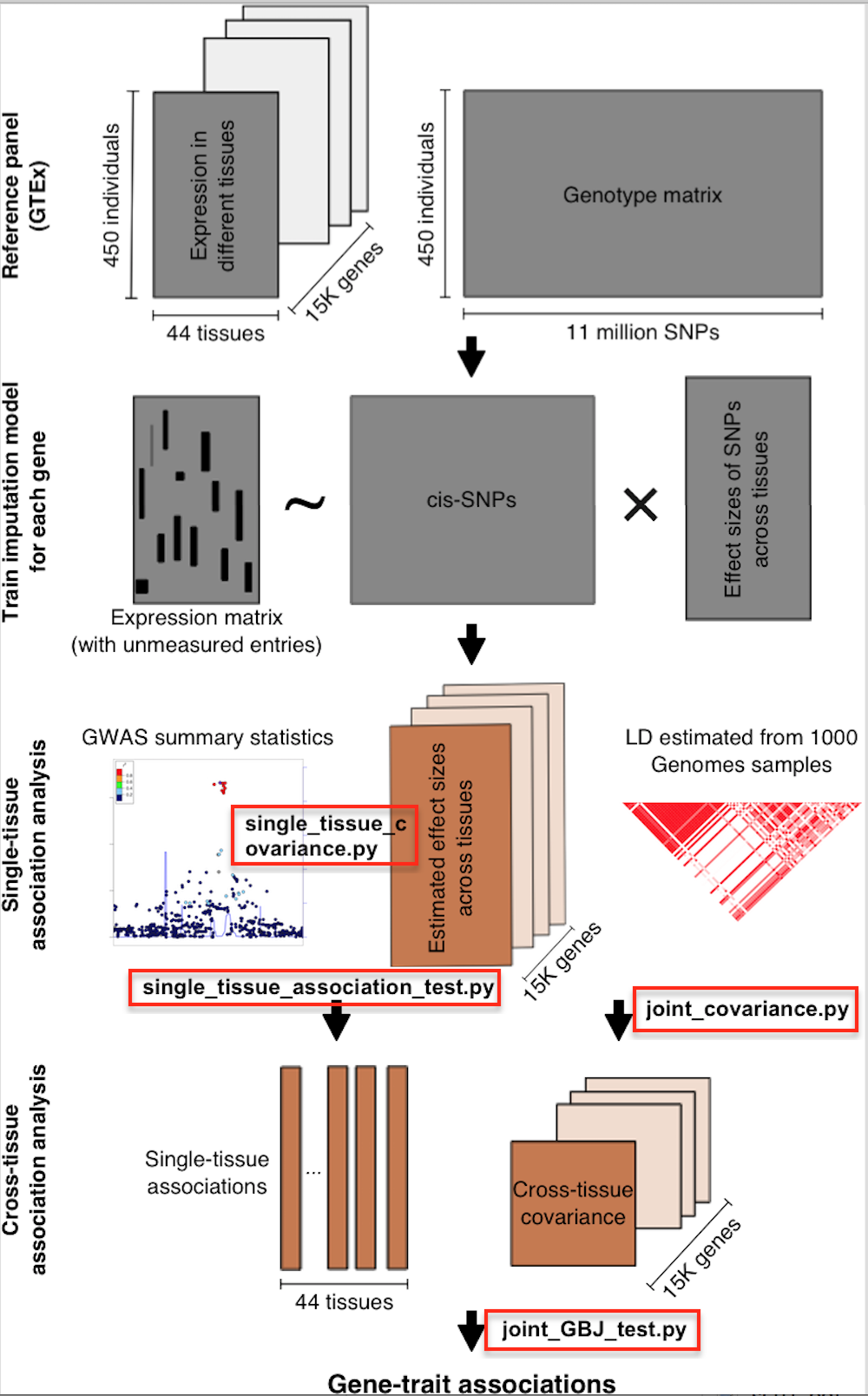
UTMOST (Unified Test for MOlecular SignaTures) is a principled method to perform cross-tissue expression imputation and gene-level association analysis. The preprint could be found at A statistical framework for cross-tissue transcriptome-wide association analysis.
The software is developed and tested in Linux and Mac OS environments.
-
Python 2.7
-
numpy (>=1.11.1)
-
scipy (>=0.18.1)
-
pandas (>=0.18.1)
-
rpy2 (==2.8.6)
-
R is needed for GBJ testing.
-
GBJ (0.5.0)
## Install python module with pip
$ pip install numpy --user
$ pip install scipy --user
$ pip install pandas --user
$ pip install -Iv rpy2==2.8.6 --user
## GBJ could be installed with R interface
install.packages('GBJ')-
single_tissue_covariance.py
-
single_tissue_association_test.py
-
joint_covariance.py
-
joint_GBJ_test.py
-
test_tool
-
metax module
The following example assumes that you have python 2.7, numpy, pandas, scipy, rpy2, R and GBJ installed. All of these functions take different number of command line parameters. Run them with --help or -h option to see the options. Codes for training cross-tissue gene-expression imputation models were curated in a separate repo.
This section is a demonstration of applying UTMOST with imputation models jointly trained in 44 tissues with GTEx data. The sample_data.zip contains pre-calculated imputation models, covariance matrices for single tissue and joint tissue GBJ test. Pipeline for generating covariance matrices with your own imputation models and incorporating other eQTL/sQTL data (e.g. from STARNET and BLUEPRINT (ftp://ftp.ebi.ac.uk/pub/databases/blueprint/blueprint_Epivar/qtl_as/)), i.e. the analysis pipeline used in manuscript) could be found in the following section.
1. Clone the UTMOST repository
$ git clone https://github.com/Joker-Jerome/UTMOST2. Go to the software directory
$ cd ./UTMOST3.1 Download imputation model (weights) data (1.9GB for zipped file, 3.4GB after unzipping)
$ wget --load-cookies /tmp/cookies.txt "https://drive.google.com/uc?export=download&confirm=$(wget --quiet --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://drive.google.com/uc?export=download&id=1u8CRwb6rZ-gSPl89qm3tKpJArUT8XrEe' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\1\n/p')&id=1u8CRwb6rZ-gSPl89qm3tKpJArUT8XrEe" -O sample_data.zip && rm -rf /tmp/cookies.txt
$ unzip sample_data.zipThis folder will include the following files/folders:
weight_db_GTEx/ ## jointly trained imputation models for 44 GTEx tissues
weight_db_external/ ## imputation models for STARNET liver tissue and BLUEPRINT 3 cell-type eQTL/sQTL data
dosage/ ## a reference genotype panel for calculating covariance matrices
GWAS/ ## a simulated GWAS summary stats file as an example
covariance.txt.gz and DGN-WB_0.5.db ## toy example for demonstrating single-tissue testTo run single-tissue and joint GBJ test with these imputation models, you need to either generate covariance matrices with a reference genotype panel (for details see Methods section in manuscript) or you could download the pre-calculated covariance matrices for 44 GTEx tissues. Instructions on how to calculate covariance matrices could be found in Section 5 in this tutorial.
3.2 Download pre-calculate covariance matrices for single-tissue/joint test (large file 28GB for zipped file, 45GB after unzipping)
- [Update]--2023/06/09--for weights trained with GTEx v8, please visit https://zhaocenter.org/UTMOST.
$ cd sample_data
$ wget --load-cookies /tmp/cookies.txt "https://drive.google.com/uc?export=download&confirm=$(wget --quiet --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://drive.google.com/uc?export=download&id=1Kh3lHyTioKIXqCsREmsAyC-dS49KVO9G' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\1\n/p')&id=1Kh3lHyTioKIXqCsREmsAyC-dS49KVO9G" -O covariance_tissue.tar.gz && rm -rf /tmp/cookies.txt
$ wget --load-cookies /tmp/cookies.txt "https://drive.google.com/uc?export=download&confirm=$(wget --quiet --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://drive.google.com/uc?export=download&id=1tqIW5Ms8p1StX7WXXWVa4TGKb5q58TPA' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\1\n/p')&id=1tqIW5Ms8p1StX7WXXWVa4TGKb5q58TPA" -O covariance_joint.zip && rm -rf /tmp/cookies.txt
$ tar -zxvf covariance_tissue.tar.gz
$ unzip covariance_joint.zip
covariance_tissue/ and covariance_joint/ contain covariance matrices required for single-tissue and joint gene-trait association tests, respectively.
3.3 Download example GWAS summary statistics GIANT GWAS Anthropometric 2015 BMI data
cd GWAS
wget https://portals.broadinstitute.org/collaboration/giant/images/1/15/SNP_gwas_mc_merge_nogc.tbl.uniq.gz
gunzip SNP_gwas_mc_merge_nogc.tbl.uniq.gz4. Run UTMOST with cross-tissue imputation models trained in 44 GTEx tissues
4.1. Run single tissue association test for 44 tissues
cd ../.. ## at UTMOST/
mkdir sample_data/results
TISSUE_GTEx=(Adipose_Subcutaneous Adipose_Visceral_Omentum Adrenal_Gland Artery_Aorta Artery_Coronary Artery_Tibial Brain_Anterior_cingulate_cortex_BA24 Brain_Caudate_basal_ganglia Brain_Cerebellar_Hemisphere Brain_Cerebellum Brain_Cortex Brain_Frontal_Cortex_BA9 Brain_Hippocampus Brain_Hypothalamus Brain_Nucleus_accumbens_basal_ganglia Brain_Putamen_basal_ganglia Breast_Mammary_Tissue Cells_EBV-transformed_lymphocytes Cells_Transformed_fibroblasts Colon_Sigmoid Colon_Transverse Esophagus_Gastroesophageal_Junction Esophagus_Mucosa Esophagus_Muscularis Heart_Atrial_Appendage Heart_Left_Ventricle Liver Lung Muscle_Skeletal Nerve_Tibial Ovary Pancreas Pituitary Prostate Skin_Not_Sun_Exposed_Suprapubic Skin_Sun_Exposed_Lower_leg Small_Intestine_Terminal_Ileum Spleen Stomach Testis Thyroid Uterus Vagina Whole_Blood)
for tissue in ${TISSUE_GTEx[@]}
do
python2 ./single_tissue_association_test.py \
--model_db_path sample_data/weight_db_GTEx/${tissue}.db \
--covariance sample_data/covariance_tissue/${tissue}.txt.gz \
--gwas_folder sample_data/GWAS \
--gwas_file_pattern SNP_gwas_mc_merge_nogc.tbl.uniq \
--snp_column SNP \
--effect_allele_column A1 \
--non_effect_allele_column A2 \
--beta_column b \
--pvalue_column p \
--output_file sample_data/results/${tissue}.csv
doneThe example command parameters:
-
--model_db_path
Path to gene expression imputation model (estimated weights/effect sizes of cis-eQTLs).
-
--covariance
Path to file containing covariance information (used to estimate the variance of gene-level effect size estimator, see Gene-level association test in Methods section of manuscript for details).
-
--gwas_folder
Folder containing GWAS summary statistics data.
-
--gwas_file_pattern
The file patten of gwas file (file name of summary statistics if not segmented by chromosomes).
-
--snp_column
Argument with the name of the column containing the RSIDs.
-
--effect_allele_column
Argument with the name of the column containing the effect allele.
-
--non_effect_allele_column
Argument with the name of the column containing the non-effect allele.
-
--beta_column
The column containing -effect size estimator for each SNP- in the input GWAS files.
-
--pvalue_column
The column containing -PValue for each SNP- in the input GWAS files.
-
--output_file
Path where results will be saved to.
4.2. Combine gene-trait associations in 44 tissues by joint GBJ test
mkdir sample_data/results_GTEx ## save association results for cross-tissue joint test
UTMOST_path=/absolute/path/to/UTMOST/
$ python2 joint_GBJ_test.py \
--weight_db $UTMOST_path/sample_data/weight_db_GTEx/ \
--output_dir $UTMOST_path/sample_data/results_GTEx/ \
--cov_dir $UTMOST_path/sample_data/covariance_joint/ \
--input_folder $UTMOST_path/sample_data/results/ \
--gene_info $UTMOST_path/intermediate/gene_info.txt \
--output_name GIANT_BMI_2015_GTEx_44_joint \
--start_gene_index 1 \
--end_gene_index 17290The example command parameters:
-
--verbosity
Log verbosity level. 1 means everything will be logged. 10 means high level messages will be logged.
-
--weight_db
Name of weight db in data folder (imputation models).
-
--input_folder
Name of folder containing single-tissue association results (generated in Section 4.1).
-
--cov_dir
Path where covariance results are (covariance matrix for gene-level test statistics across tissues, see Gene-level association test in Methods section of manuscript for details).
-
--output_dir
Path where results will be saved to.
-
--gene_info
File containing the all the genes tested.
-
--start_gene_index
Index of the starting gene in intermediate/gene_info.txt (for parallel computing purpose, could test multiple gene at the same time to reduce computation time).
-
--end_gene_index
Index of the ending gene in intermediate/gene_info.txt (for parallel computing purpose, could test multiple gene at the same time to reduce computation time).
Output format:
| Gene | Test score | P value |
|---|---|---|
| Gene A | test score A | P value A |
| Gene B | test score B | P value B |
Using STARNET and BLUEPRINT (ftp://ftp.ebi.ac.uk/pub/databases/blueprint/blueprint_Epivar/qtl_as/) as an example, for details, please see Results and Methods sections of manuscript
Note: this part also requires data in sample_data folder
5.1. Calculate the single tissue covariance
TISSUE_external=(Liver_STARNET1 mono_eqtl mono_sqtl neut_eqtl neut_sqtl tcel_eqtl tcel_sqtl)
mkdir sample_data/covariance_external
for tissue in ${TISSUE_external[@]}
do
python2 ./single_tissue_covariance.py \
--weight_db sample_data/weight_db_external/${tissue}.db \
--input_folder sample_data/dosage/ \
--covariance_output sample_data/covariance_external/${tissue}.txt.gz
doneThe example command parameters:
-
--weight_db
Path to tissue transriptome model.
-
--input_folder
Folder containing GWAS summary statistics data.
-
--covariance_output
Path where covariance will be saved to.
5.2. Run the single tissue association test
for tissue in ${TISSUE_external[@]}
do
python2 ./single_tissue_association_test.py \
--model_db_path sample_data/weight_db_external/${tissue}.db \
--covariance sample_data/covariance_external/${tissue}.txt.gz \
--gwas_folder sample_data/GWAS \
--gwas_file_pattern SNP_gwas_mc_merge_nogc.tbl.uniq \
--snp_column SNP \
--effect_allele_column A1 \
--non_effect_allele_column A2 \
--beta_column b \
--pvalue_column p \
--output_file sample_data/results/${tissue}.csv
done5.3. Calculate the joint tissue covariance
mkdir covariance_GTEx_external ## path for saving new covariance matrix (could take ~25GB space)
mkdir sample_data/weight_db_GTEx_external ## path for saving imputation models across different tissues
cp sample_data/weight_db_GTEx/* sample_data/weight_db_GTEx_external/
cp sample_data/weight_db_external/* sample_data/weight_db_GTEx_external/
python2 ./joint_covariance.py \
--weight_db sample_data/weight_db_GTEx_external/ \
--input_folder sample_data/dosage/ \
--covariance_output sample_data/covariance_GTEx_external/The example command parameters:
-
--verbosity
Log verbosity level. 1 means everything will be logged. 10 means high level messages will be logged.
-
--weight_db
Name of weight db in data folder.
-
--input_folder
Name of folder containing dosage data.
-
--covariance_output
Path where covariance results will be saved to.
-
--min_maf_filter
Filter SNPs according to this maf.
-
--max_maf_filter
Filter SNPs according to this maf.
5.4. Combine gene-trait associations in 44 tissues + STARNET liver eQTL + BLUEPRINT eQTL/sQTL by joint GBJ test
## note after 5.2, sample_data/results/ now contains 44 + 1 + 3*2 single-tissue association results
UTMOST_path=/absolute/path/to/UTMOST/
$ mkdir results_GTEx_external
$ python2 joint_GBJ_test.py \
--weight_db $UTMOST_path/sample_data/weight_db_GTEx_external/ \
--output_dir $UTMOST_path/results_GTEx_external/ \
--cov_dir $UTMOST_path/covariance_GTEx_external/ \
--input_folder $UTMOST_path/sample_data/results/ \
--gene_info $UTMOST_path/intermediate/gene_info.txt \
--output_name test_GTEx_externalConditional Analysis
python2 conditional_test_geneset.py \
--utmost_dir $UTMOST_path \
--weight_db $UTMOST_path/sample_data/weight_db_GTEx/ \
--input_folder $UTMOST_path/sample_data/dosage/ \
--gwas_str gwas \
--gwas_folder /GWAS_data/ \
--gwas_file_pattern GWAS.txt \
--snp_column SNP \
--effect_allele_column A1 \
--non_effect_allele_column A2 \
--beta_column BETA \
--se_column SE \
--pvalue_column P \
--chr_idx 1 \
--gene_list Gene1,Gene2,Gene3 \
--output_dir /output_dir/The example command parameters:
-
--utmost_dir
Path to UTMOST.
-
--weight_db
Path to gene expression imputation model (estimated weights/effect sizes of cis-eQTLs).
-
--input_folder
Name of folder containing dosage data.
-
--gwas_str
GWAS name string.
-
--gwas_folder
Folder containing GWAS summary statistics data.
-
--gwas_file_pattern
The file patten of gwas file (file name of summary statistics if not segmented by chromosomes).
-
--snp_column
Argument with the name of the column containing the RSIDs.
-
--effect_allele_column
Argument with the name of the column containing the effect allele.
-
--non_effect_allele_column
Argument with the name of the column containing the non-effect allele.
-
--beta_column
The column containing -effect size estimator for each SNP- in the input GWAS files.
-
--se_column
The column containing the standard error for effect size estimate.
-
--pvalue_column
The column containing -PValue for each SNP- in the input GWAS files.
-
--chr_idx
Chromosome number.
-
--gene_list
Genes to be tested.
-
--output_dir
Path where results will be saved to.
Part of the code is modified from MetaXcan https://github.com/hakyimlab/MetaXcan. We thank the authors for sharing the code.
Hu et al. (2018). A statistical framework for cross-tissue transcriptome-wide association analysis. bioRxiv, 286013. Link