GeneidX provides a fast annotation of the protein-coding genes in an eukaryotic genome taking as input the genome assembly and the taxonomic ID of the species to annotate.
In the description here, you can find our preliminary results, a schema of our method and a description of the minimal requirements and commands required for running it.
Stay tuned for an article with detailed descriptions and feel free to contact us if you are trying it and find any problem.
The results of an initial benchmarking using vertebrates genomes annotated in Ensembl show that our method is as accurate as the top ab initio gene predictors, but it is between 10 and 100 times faster.
Having defined the parameters and ensuring that Nextflow and a container technology are installed.
This pipeline requires the compressed genome assembly and the taxid of the species to annotate.
export NXF_VER=22.04.4
nextflow run guigolab/geneidx -profile <docker/singularity>
--assembly <GENOME>.fa,gz
--taxid <TAXID>
--outdir <OUTPUT_directory>
or alternatively, clone the repository and then run it (highly recommended)
git clone https://github.com/guigolab/geneidx.git
cd geneidx
export NXF_VER=22.04.4
nextflow run main.nf -profile <docker/singularity>
--assembly <GENOME>.fa.gz
--taxid <TAXID>
--outdir <OUTPUT_directory>
Revise the DETAILS section below for the minor specifications of each parameter.
Geneidx is working in Nextflow version 22.04.4 although we are working on updating the code up to make it work with the latest versions.
- Make sure ( Docker or Singularity ) and Nextflow are installed in your computer.
- Define the input parameters:
- Compressed FASTA file with
.fa.gztermination. - Taxid of the species to annotate or from the closest relative described with taxid. You can find this information in NCBI Taxonomy database.
- Optional parameters:
- Proteins from closely related species: (choose one of these options)
- Provide a compressed FASTA file with the proteins selected.
- Let the pipeline download the proteins automatically from UniRef90 (nothing should be provided in this case.)
- Tune parameters for the gene prediction process.
- Indicate the values for each Geneid parameter.
- Let the pipeline select them from the closest pretrained Geneid parameter file. (find the parameters in data/Parameter_files.taxid/)
- Find more information here: GENEID parameter files
Which steps are taking place as part of this pipeline?
This is a graphical summary, and the specific steps are outlined below.
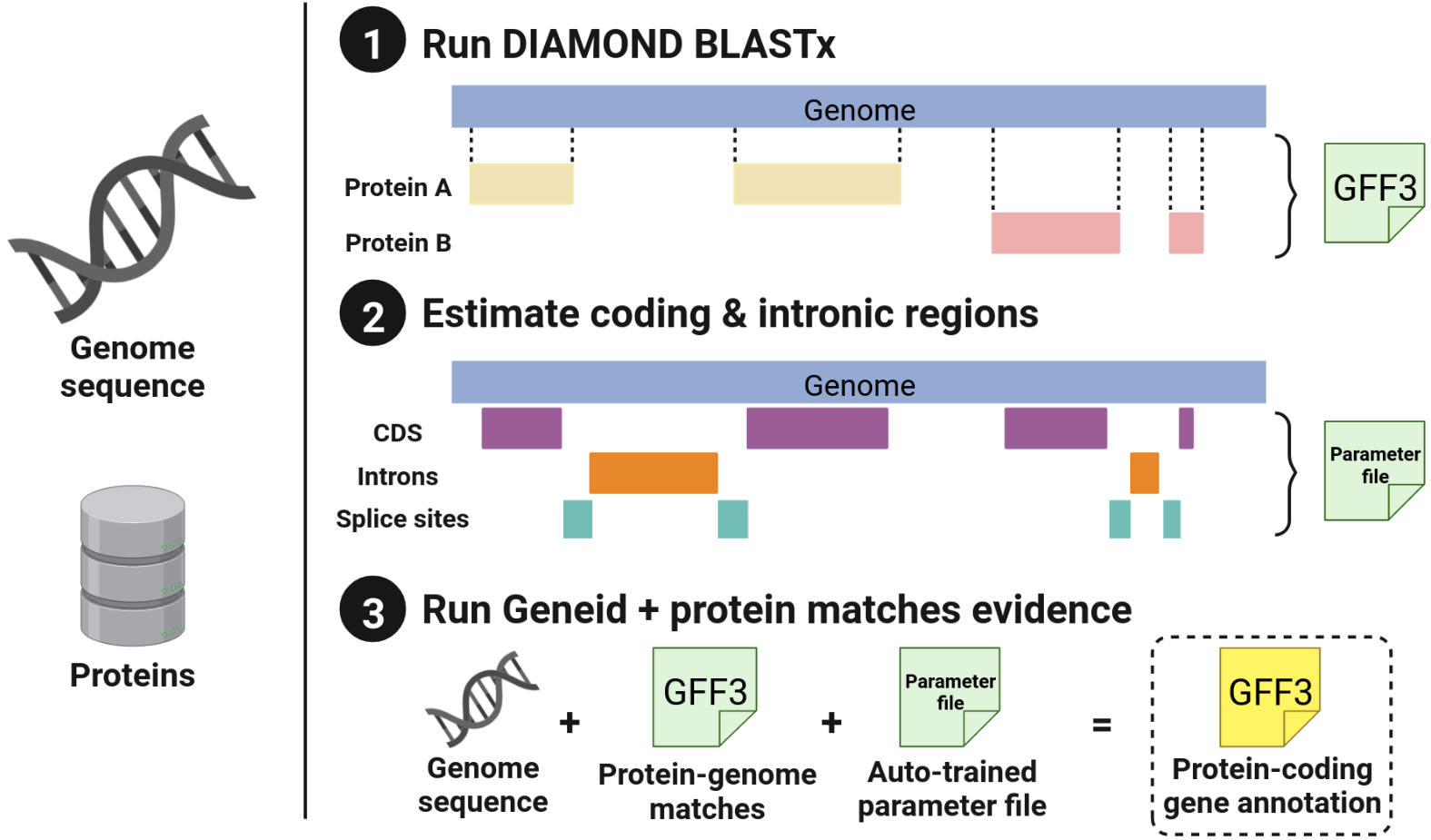
- Get the set of proteins to be used for the protein-to-genome alignments.
- Get the closest Geneid parameter file to use, as source of the parameters is not indicated by the user.
- Create the protein database for DIAMOND to use it as a source.
- Align the provided genome against the database created using DIAMOND BLASTx flavour.
- Run the auto-training process:
- Use matches to estimate the coding sections, look for open reading frames between stop codons.
- Use matches from the same protein to predict the potential introns.
- From the sequences of both previous steps compute the initial and transition probability matrices required for the computation of the coding potential of the genome that will be annotated.
- Update the parameter file with the parameters indicated in the params.config file and also the matrices automatically generated from the protein-DNA matches.
- Run Geneid with the new parameter file and the protein-DNA matches from the previous steps as additional evidence. This is done in parallel for each independent sequence inside the genome FASTA file, and it consists of the following steps:
- Pre-process the matches to join those that overlap and re-score them appropriately.
- Run Geneid using the evidence and obtain the GFF3 file.
- Remove the files from the internal steps.
- Concatenate all the outputs into a single GFF3 file that is sorted by coordinates.
- Add information from proteins matching the predicted genes.
-
The name of the sequences in the FASTA file cannot contain unusual characters.
-
The input genome file must be a gzip-compressed FASTA file. (.fa.gz)
-
Auto-train the parameter file always.
-
It is recommended to clone the repository and then run the pipeline from there.
-
So far the output of the predictions is not stored in the path indicated when running the pipeline, but in output/species/{taxid of the species}.
-
If you are running the pipeline multiple times, it is recommended that you define a directory for downloading the docker/singularity images to avoid having to download them multiple times. See
singularity.cacheDir variableinnextflow.config. -
If you are starting from an unmasked genome assembly, check another version of this pipeline integrating the genome masking process. GeneidXmask repository.
-
If you have used Geneid in the past and have manually trained a parameter file, we are open to receive them and share them in our repositories giving credit to the users who generated them. Contact us at ferriol.calvet@crg.eu.
Follow us on Twitter (@GuigoLab) for updates in the article describing our method, and contact us for any questions or suggestions.