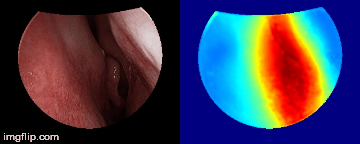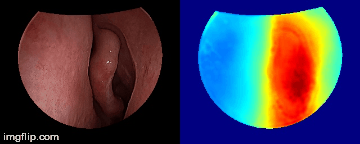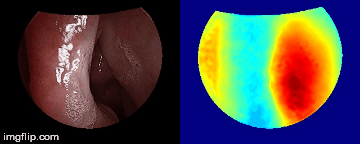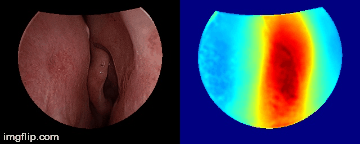
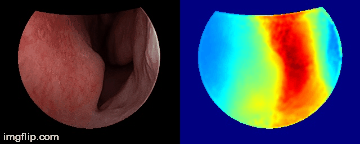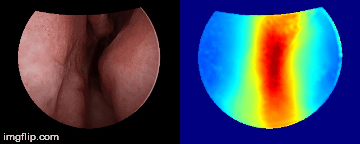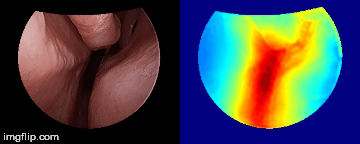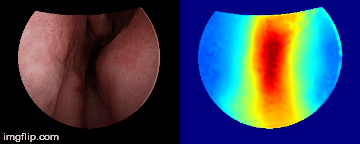
Depth predictions come from a testing patient and camera which are not seen during training. Depth maps are normalized per frame for display purpose.
This codebase implements the system described in the paper:
Self-supervised Learning for Dense Depth Estimation in Monocular Endoscopy
Xingtong Liu, Ayushi Sinha, Mathias Unberath, Masaru Ishii, Gregory D. Hager, Russell H. Taylor, Austin Reiter
In International Workshop on Computer-Assisted and Robotic Endoscopy 2018 (Best paper award & Best presentation award)
The journal version of this paper has been submitted to IEEE Transactions on Medical Imaging under review.
Please contact Xingtong Liu (xliu89@jh.edu) if you have any questions.
Please cite CARE Workshop or TMI Submission Arxiv Preprint if you use the code here.
@incollection{liu2018self,
title={Self-supervised Learning for Dense Depth Estimation in Monocular Endoscopy},
author={Liu, Xingtong and Sinha, Ayushi and Unberath, Mathias and Ishii, Masaru and Hager, Gregory D and Taylor, Russell H and Reiter, Austin},
booktitle={OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis},
pages={128--138},
year={2018},
publisher={Springer}
}
@ARTICLE{2019arXiv190207766L,
author = {{Liu}, Xingtong and {Sinha}, Ayushi and {Ishii}, Masaru and
{Hager}, Gregory D. and {Reiter}, Austin and {Taylor}, Russell H. and
{Unberath}, Mathias},
title = "{Self-supervised Learning for Dense Depth Estimation in Monocular Endoscopy}",
journal = {arXiv e-prints},
keywords = {Computer Science - Computer Vision and Pattern Recognition, Statistics - Machine Learning},
year = "2019",
month = "Feb",
eid = {arXiv:1902.07766},
pages = {arXiv:1902.07766},
archivePrefix = {arXiv},
eprint = {1902.07766},
primaryClass = {cs.CV},
adsurl = {https://ui.adsabs.harvard.edu/\#abs/2019arXiv190207766L},
adsnote = {Provided by the SAO/NASA Astrophysics Data System}
}
-
Install all necessary python packages: Pytorch, OpenCV, numpy, tqdm, pathlib, torchsummary, tensorboardX, albumentations, argparse, pickle, plyfile, yaml, datetime, shutil, matplotlib
-
Generate training data from training videos using Structure from Motion (SfM) or Simultaneous Localization and Mapping (SLAM). Refer to one training example in this repository.
-
Run teacher_training.py with proper arguments for teacher-self-supervised learning. One example is:
/path/to/python /path/to/teacher_training.py --adjacent_range 10 20 --max_lr 1.0e-3 --min_lr 1.0e-4 --testing_patient_id 2 --use_hsv_colorspace --load_intermediate_data --number_epoch 4 --ssl_weight 0.3 --dcl_weight 10.0 --sfl_weight 100.0 --batch_size 8 --num_workers 8 --training_data_path "/path/to/training/data" --training_root "/path/to/training/directory"
- Run teacher_student_training.py with proper arguments for teacher-supervise-student learning. One example is:
/path/to/python /path/to/teacher_student_training.py --adjacent_range 10 20 --max_lr 1.0e-3 --min_lr 1.0e-4 --testing_patient_id 2 --use_hsv_colorspace --load_intermediate_data --number_epoch 1 --batch_size 8 --num_workers 8 --training_data_path "/path/to/training/data" --training_root "/path/to/training/directory"
- Run teacher_student_training.py with proper arguments for student-self-supervised learning. One example is:
/path/to/python /path/to/teacher_student_training.py --adjacent_range 10 20 --max_lr 1.0e-3 --min_lr 1.0e-4 --testing_patient_id 2 --student_learn_from_sfm --load_intermediate_data --use_hsv_colorspace --number_epoch 12 --ssl_weight 0.3 --dcl_weight 10.0 --sfl_weight 100.0 --batch_size 8 --num_workers 4 --use_previous_student_model --training_data_path "/path/to/training/data" --training_root "/path/to/training/directory"
- Run evaluate.py to generate evaluation results. Apply any rigid registration algorithm to register the predicted point clouds to the corresponding CT model to calculate residual errors (this step may require manual point cloud initialization).
This code is only experimental and not ready for clinical applications.
Authors are not responsible for any accidents related to this repository.
This code is only allowed for non-commercial usage.