By Louie Letourneau, MSc.
Edited and Modified by Mathieu Bourgey, Ph.D and Robert Eveleigh, MSc.
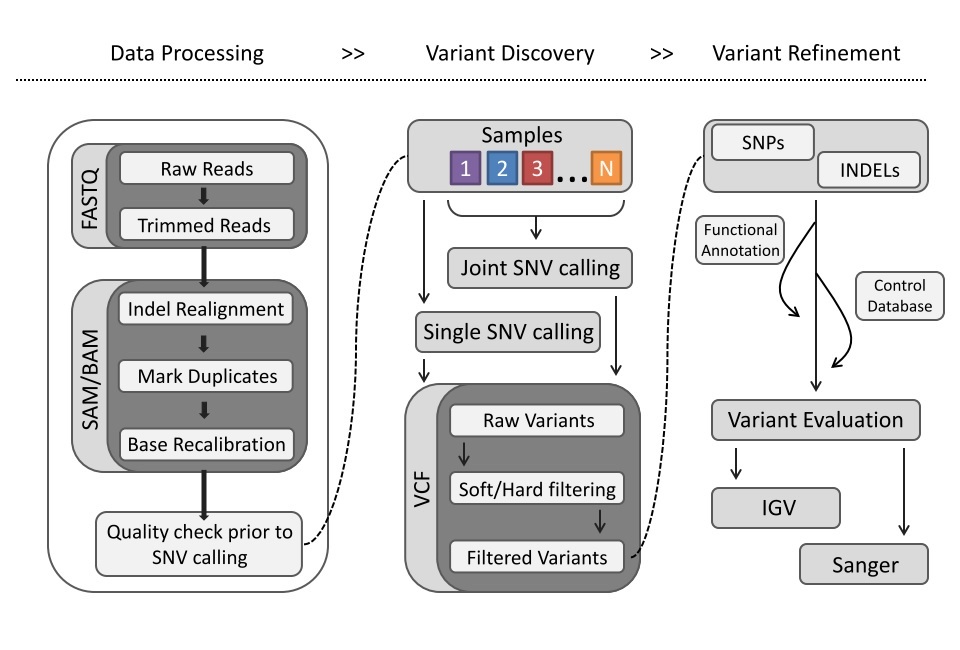
In this workshop, we will present the main steps that are commonly used to process and to analyze sequencing data. We will focus only on whole genome data and provide command lines that allow detecting Single Nucleotide Variants (SNV). This workshop will show you how to launch individual steps of a complete DNA-Seq pipeline
We will be working on a 1000 genome sample, NA12878. You can find the whole raw data on the 1000 genome website
For practical reasons we subsampled the reads from the sample because running the whole dataset would take way too much time and resources.
This work is licensed under a Creative Commons Attribution-ShareAlike 3.0 Unported License. This means that you are able to copy, share and modify the work, as long as the result is distributed under the same license.
- You can find all the unix command lines of this practical in the file: commands.sh
export PICARD_JAR=/usr/local/bin/picard.jar
export GATK_JAR=/usr/local/bin/GenomeAnalysisTK.jar
export BVATOOLS_JAR=/usr/local/bin/bvatools-1.6-full.jar
export TRIMMOMATIC_JAR=/usr/local/bin/trimmomatic-0.35.jar
export REF=/home/mBourgey/kyoto_workshop_WGS_2015/references/
cd $HOME
rsync -avP /home/reveleigh/cleanCopy/workshop $HOME
cd $HOME/workshop/
ls
The initial structure of your folders should look like this:
<ROOT>
|-- raw_reads/ # fastqs from the center (down sampled)
`-- NA12878 # One sample directory
|-- runERR_1 # Lane directory by run number. Contains the fastqs
`-- runSRR_1 # Lane directory by run number. Contains the fastqs
These are all already installed, but here are the original links.
So you've just received an email saying that your data is ready for download from the sequencing center of your choice.
What should you do? solution
Let's first explore the fastq file.
Try these commands
zless -S raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair1.fastq.gz
Why was it like that ? solution
Now try these commands:
zcat raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair1.fastq.gz | head -n4
zcat raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair2.fastq.gz | head -n4
What was special about the output ?
Why was it like that? Solution
You could also just count the reads
zgrep -c "^@SRR" raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair1.fastq.gz
We should obtain 15546 reads
Why shouldn't you just do ?
zgrep -c "^@" raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair1.fastq.gz
####Phred Score In the original implementation of Phred scores, information about the size and shape of peaks in sequencing trace files were used to generate an error probability and converted to a log score.
The same principles are used for base quality made by the base calling method of the NGS sequencer i.e the base quality is the probability of the base to have been called incorrectly
Phred score/Base quality = - 10 * log10(error probability)
e.g. Base Quality of 10
10 = -10 * log10(error probability)
-1 = log10(error probability)
10^-1 = error probability *this means 10 to the -1 power
1 / 10 = error probability
| Phred Quality Score | Probability of incorrect base call | Base call accuracy |
|---|---|---|
| 10 | 1 in 10 | 90% |
| 20 | 1 in 100 | 99% |
| 30 | 1 in 1000 | 99.9% |
| 40 | 1 in 10,000 | 99.99% |
| 50 | 1 in 100,000 | 99.999% |
| 60 | 1 in 1,000,000 | 99.9999% |
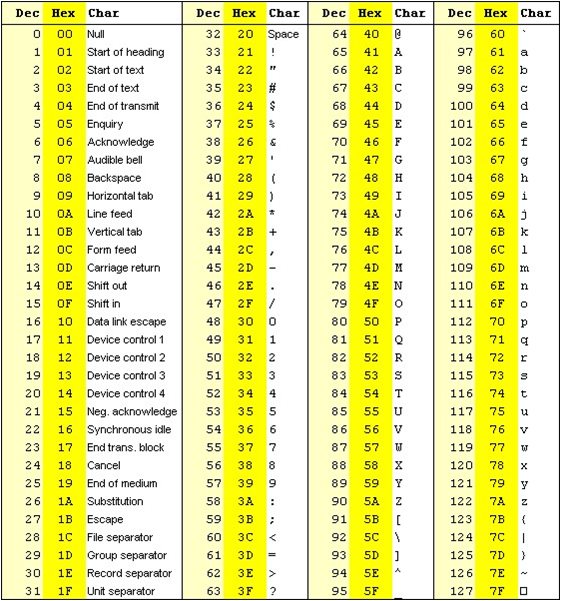
The base quality score however is respresented as a ASCII code in fastq and other file formats
Typically the phred33 (sanger format) scale is used by adding 33 to the Phred quality score and looking up the corresponding ASCII character
Example
Phred score of 31
31+33 = 64
In Dec column 64 corresponds to '@' ASCII character
See below
We can't look at all the reads. Especially when working with whole genome 30x data. You could easily have billions of reads.
Tools like FastQC and BVATools readsqc can be used to plot many metrics from these data sets.
Let's look at the data:
Try commands:
mkdir originalQC/
java -Xmx1G -jar ${BVATOOLS_JAR} readsqc \
--read1 raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair1.fastq.gz \
--read2 raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair2.fastq.gz \
--threads 2 --regionName SRR --output originalQC/
java -Xmx1G -jar ${BVATOOLS_JAR} readsqc \
--read1 raw_reads/NA12878/runERR_1/NA12878.ERR.33.pair1.fastq.gz \
--read2 raw_reads/NA12878/runERR_1/NA12878.ERR.33.pair2.fastq.gz \
--threads 2 --regionName ERR --output originalQC/
Copy the images from the originalQC folder to your desktop and open the images.
scp -r <USER>@www.genome.med.kyoto-u.ac.jp:~/workshop/originalQC/ ./
Open the images
What stands out in the graphs ? Solution
All the generated graphics have their uses. Two particularly useful to get an overall picture of sequencing quality.
- The quality box plots - Box whisker type plots showing the quality distribution of your data
- The nucleotide content graphs - Plots the proportion of each base position in a file
General observations:
- Some reads have bad 3' ends.
- Some reads have adapter sequences in them.
Trimming of the reads is required to remove the low quality bases at the 3' end, include reads of a specific minimum length and trim adapters and other technical sequences
To do that we will use Trimmomatic using these options:
- ILLUMINACLIP: Cut adapter and other illumina-specific sequences from the read (2:30:15)
- TRAILING: Cut bases off the end of a read, if below a threshold quality (20)
- MINLEN: Drop the read if it is below a specified length (32)
- TOPHRED33: Convert quality scores to Phred-33
The adapter file is in your work folder.
cat adapters.fa
Why are there 2 different ones ? Solution
Try command:
mkdir -p reads/NA12878/runSRR_1/
mkdir -p reads/NA12878/runERR_1/
java -Xmx2G -cp $TRIMMOMATIC_JAR org.usadellab.trimmomatic.TrimmomaticPE -threads 2 -phred33 \
raw_reads/NA12878/runERR_1/NA12878.ERR.33.pair1.fastq.gz \
raw_reads/NA12878/runERR_1/NA12878.ERR.33.pair2.fastq.gz \
reads/NA12878/runERR_1/NA12878.ERR.t20l32.pair1.fastq.gz \
reads/NA12878/runERR_1/NA12878.ERR.t20l32.single1.fastq.gz \
reads/NA12878/runERR_1/NA12878.ERR.t20l32.pair2.fastq.gz \
reads/NA12878/runERR_1/NA12878.ERR.t20l32.single2.fastq.gz \
ILLUMINACLIP:adapters.fa:2:30:15 TRAILING:20 MINLEN:32 \
2> reads/NA12878/runERR_1/NA12878.ERR.trim.out
java -Xmx2G -cp $TRIMMOMATIC_JAR org.usadellab.trimmomatic.TrimmomaticPE -threads 2 -phred33 \
raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair1.fastq.gz \
raw_reads/NA12878/runSRR_1/NA12878.SRR.33.pair2.fastq.gz \
reads/NA12878/runSRR_1/NA12878.SRR.t20l32.pair1.fastq.gz \
reads/NA12878/runSRR_1/NA12878.SRR.t20l32.single1.fastq.gz \
reads/NA12878/runSRR_1/NA12878.SRR.t20l32.pair2.fastq.gz \
reads/NA12878/runSRR_1/NA12878.SRR.t20l32.single2.fastq.gz \
ILLUMINACLIP:adapters.fa:2:30:15 TRAILING:20 MINLEN:32 \
2> reads/NA12878/runSRR_1/NA12878.SRR.trim.out
cat reads/NA12878/runERR_1/NA12878.ERR.trim.out reads/NA12878/runSRR_1/NA12878.SRR.trim.out
What does Trimmomatic says it did ? Solution
Let's look at the graphs now
Try command:
mkdir postTrimQC/
java -Xmx1G -jar ${BVATOOLS_JAR} readsqc \
--read1 reads/NA12878/runERR_1/NA12878.ERR.t20l32.pair1.fastq.gz \
--read2 reads/NA12878/runERR_1/NA12878.ERR.t20l32.pair2.fastq.gz \
--threads 2 --regionName ERR --output postTrimQC/
java -Xmx1G -jar ${BVATOOLS_JAR} readsqc \
--read1 reads/NA12878/runSRR_1/NA12878.SRR.t20l32.pair1.fastq.gz \
--read2 reads/NA12878/runSRR_1/NA12878.SRR.t20l32.pair2.fastq.gz \
--threads 2 --regionName SRR --output postTrimQC/
How does it look now ? Solution
The raw reads are now cleaned up of artefacts we can align each lane separately.
Why should this be done separatly? Solution
Why is it important to set Read Group information ? Solution
##Alignment with bwa-mem
Try command:
mkdir -p alignment/NA12878/runERR_1
mkdir -p alignment/NA12878/runSRR_1
bwa mem -M -t 2 \
-R '@RG\tID:ERR_ERR_1\tSM:NA12878\tLB:ERR\tPU:runERR_1\tCN:Broad Institute\tPL:ILLUMINA' \
${REF}/b37.fasta \
reads/NA12878/runERR_1/NA12878.ERR.t20l32.pair1.fastq.gz \
reads/NA12878/runERR_1/NA12878.ERR.t20l32.pair2.fastq.gz \
| java -Xmx2G -jar ${PICARD_JAR} SortSam \
INPUT=/dev/stdin \
OUTPUT=alignment/NA12878/runERR_1/NA12878.ERR.sorted.bam \
CREATE_INDEX=true VALIDATION_STRINGENCY=SILENT SORT_ORDER=coordinate MAX_RECORDS_IN_RAM=500000
bwa mem -M -t 2 \
-R '@RG\tID:SRR_SRR_1\tSM:NA12878\tLB:SRR\tPU:runSRR_1\tCN:Broad Institute\tPL:ILLUMINA' \
${REF}/b37.fasta \
reads/NA12878/runSRR_1/NA12878.SRR.t20l32.pair1.fastq.gz \
reads/NA12878/runSRR_1/NA12878.SRR.t20l32.pair2.fastq.gz \
| java -Xmx2G -jar ${PICARD_JAR} SortSam \
INPUT=/dev/stdin \
OUTPUT=alignment/NA12878/runSRR_1/NA12878.SRR.sorted.bam \
CREATE_INDEX=true VALIDATION_STRINGENCY=SILENT SORT_ORDER=coordinate MAX_RECORDS_IN_RAM=500000
Why did we pipe the output of one to the other ? Solution
Could we have done it differently ? Solution
We will explore the generated BAM format in the next section.
The results lane-specific bam will now to merged into one bam file for further processing.
Since we tagged each read in the BAM by read group, upon merging we can still identify the origin of each read.
Try command:
java -Xmx2G -jar ${PICARD_JAR} MergeSamFiles \
INPUT=alignment/NA12878/runERR_1/NA12878.ERR.sorted.bam \
INPUT=alignment/NA12878/runSRR_1/NA12878.SRR.sorted.bam \
OUTPUT=alignment/NA12878/NA12878.sorted.bam \
VALIDATION_STRINGENCY=SILENT CREATE_INDEX=true
You should now have one BAM containing all your data.
Let's double check
ls -l alignment/NA12878/
samtools view -H alignment/NA12878/NA12878.sorted.bam | grep "^@RG"
You should have your 2 read group entries.
Why did we use the -H switch? Solution
Try without. What happens? Solution
We began by running quality control tools on the raw reads. Additional steps are also required to fix biases introduced at various point of the data processing procedures.
##Indel Realignment Due to the heuristics of the alignment algorithm (i.e. each read pair is optimized independent of the full alignment context), mismatches can be introduced near regions of insertions and deletions (indels).
The Genome Analysis ToolKit (GATK) has a tool called IndelRealigner which performs two steps
- Realigner Target Creator : Identifies small suspicious regions requiring realignment
- Indel Realigner : Uses the full alignment context to determine if a indel exist
Try command:
java -Xmx2G -jar ${GATK_JAR} \
-T RealignerTargetCreator \
-R ${REF}/b37.fasta \
-o alignment/NA12878/realign.intervals \
-I alignment/NA12878/NA12878.sorted.bam \
-L 1
java -Xmx2G -jar ${GATK_JAR} \
-T IndelRealigner \
-R ${REF}/b37.fasta \
-targetIntervals alignment/NA12878/realign.intervals \
-o alignment/NA12878/NA12878.realigned.sorted.bam \
-I alignment/NA12878/NA12878.sorted.bam
How could we make this go faster? Solution
How many regions were identified for realignment? Solution
This step ensures that all mate-pair information is in rync between each read and its mate pair.
NOTE: This step is deprecated now since indel realignment performs this step. We shall skip this due to time constrains
This procedure removes/marks duplicate reads in your bam file. Removing/marking duplicates from an analyses can prevent skewed allele frequencies in SNP discovery, false expression profiles in RNA-seq experiments, etc.
So what are duplicate reads? Solution
What causes these type of reads? Solution
How are duplicated reads detected? Solution
Here we will use Picard approach:
Try these command:
java -Xmx2G -jar ${PICARD_JAR} MarkDuplicates \
REMOVE_DUPLICATES=false CREATE_MD5_FILE=true VALIDATION_STRINGENCY=SILENT CREATE_INDEX=true \
INPUT=alignment/NA12878/NA12878.realigned.sorted.bam \
OUTPUT=alignment/NA12878/NA12878.sorted.dup.bam \
METRICS_FILE=alignment/NA12878/NA12878.sorted.dup.metrics
We can look in the metrics output to see what happened.
less alignment/NA12878/NA12878.sorted.dup.metrics
How many duplicates were there? Solution
We can see that it computed separate measures for each library.
Why is this important to do not combine everything? Solution
GATK BaseRecalibrator: Accurately recalibrates base quality scores by correcting for various sources of systematic technical error e.g. sequence context, mismatch position within read, etc.
Why do we need to recalibrate base quality scores ? Solution
java -Xmx2G -jar ${GATK_JAR} \
-T BaseRecalibrator \
-nct 2 \
-R ${REF}/b37.fasta \
-knownSites ${REF}/dbSnp-137.vcf.gz \
-L 1:47000000-47171000 \
-o alignment/NA12878/NA12878.sorted.dup.recalibration_report.grp \
-I alignment/NA12878/NA12878.sorted.dup.bam
java -Xmx2G -jar ${GATK_JAR} \
-T PrintReads \
-nct 2 \
-R ${REF}/b37.fasta \
-BQSR alignment/NA12878/NA12878.sorted.dup.recalibration_report.grp \
-o alignment/NA12878/NA12878.sorted.dup.recal.bam \
-I alignment/NA12878/NA12878.sorted.dup.bam
Now all refinement steps of the bam are complete. Now it is best to perform a quality check of the data.
Compute coverage For Exome data the capture kit regions are used to verify how well your targets were captured
Insert Size Used to validate the quality of the library preparaion
Alignment metrics Verifies the concordance between your sample and the reference genome
Both GATK and BVATools have depth of coverage tools.
GATK DepthOfCoverage
Try command:
java -Xmx2G -jar ${GATK_JAR} \
-T DepthOfCoverage \
--omitDepthOutputAtEachBase \
--summaryCoverageThreshold 10 \
--summaryCoverageThreshold 25 \
--summaryCoverageThreshold 50 \
--summaryCoverageThreshold 100 \
--start 1 --stop 500 --nBins 499 -dt NONE \
-R ${REF}/b37.fasta \
-o alignment/NA12878/NA12878.sorted.dup.recal.coverage \
-I alignment/NA12878/NA12878.sorted.dup.recal.bam \
-L 1:47000000-47171000
note on DepthOfCoverage command
Coverage is the expected ~30x
Look at the coverage:
less -S alignment/NA12878/NA12878.sorted.dup.recal.coverage.sample_interval_summary
Is the coverage fit with the expectation ? solution
It corresponds to the size of DNA fragments sequenced.
Different from the gap size (= distance between reads) !
These metrics are computed using Picard CollectInsertSizeMetrics:
Try command:
java -Xmx2G -jar ${PICARD_JAR} CollectInsertSizeMetrics \
VALIDATION_STRINGENCY=SILENT \
REFERENCE_SEQUENCE=${REF}/b37.fasta \
INPUT=alignment/NA12878/NA12878.sorted.dup.recal.bam \
OUTPUT=alignment/NA12878/NA12878.sorted.dup.recal.metric.insertSize.tsv \
HISTOGRAM_FILE=alignment/NA12878/NA12878.sorted.dup.recal.metric.insertSize.histo.pdf \
METRIC_ACCUMULATION_LEVEL=LIBRARY
Look at the output:
less -S alignment/NA12878/NA12878.sorted.dup.recal.metric.insertSize.tsv
There is something interesting going on with our library ERR.
Can you tell what it is? Solution
For the alignment metrics, samtools flagstat is very fast but with bwa-mem since some reads get broken into pieces, the numbers are a bit confusing.
We prefer the Picard way of computing metrics:
java -Xmx2G -jar ${PICARD_JAR} CollectAlignmentSummaryMetrics \
VALIDATION_STRINGENCY=SILENT \
REFERENCE_SEQUENCE=${REF}/b37.fasta \
INPUT=alignment/NA12878/NA12878.sorted.dup.recal.bam \
OUTPUT=alignment/NA12878/NA12878.sorted.dup.recal.metric.alignment.tsv \
METRIC_ACCUMULATION_LEVEL=LIBRARY
explore the results
less -S alignment/NA12878/NA12878.sorted.dup.recal.metric.alignment.tsv
Do you think the sample and the reference genome fit together ? Solution
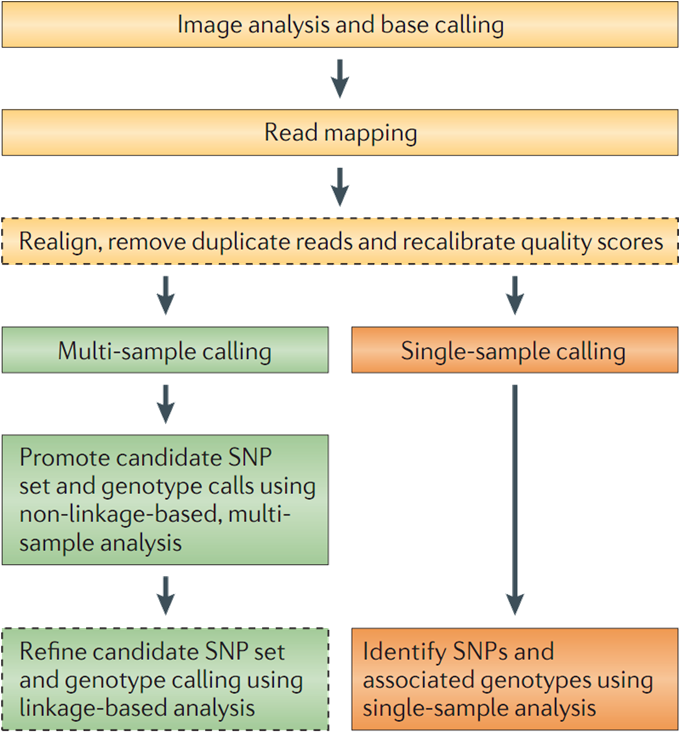
Most of SNV caller use either a Baysian, a threshold or a t-test approach to do the calling
I won't go into the details of finding which variant is good or bad since this will be your next workshop.
Here we will just call and view the variants using the samtools mpileup function:
mkdir variants
samtools mpileup -L 1000 -B -q 1 -g \
-f ${REF}/b37.fasta \
-r 1:47000000-47171000 \
alignment/NA12878/NA12878.sorted.dup.recal.bam | bcftools view -vcg - \
> variants/mpileup.vcf
note on samtools mpileup and bcftools command
Now we have variants from all three methods. Let's compress and index the vcfs for future visualisation.
bgzip -c variants/mpileup.vcf > variants/mpileup.vcf.gz
tabix -p vcf variants/mpileup.vcf.gz
Let's look at the compressed vcf.
zless -S variants/mpileup.vcf.gz
Details on the spec can be found here: http://vcftools.sourceforge.net/specs.html
Fields vary from caller to caller.
Some values are are almost always there:
- The ref vs alt alleles,
- variant quality (QUAL column)
- The per-sample genotype (GT) values.
#Add-on Additional exercice to play with sam/bam files
This tutorial is an adaptation of the one created by Louis letourneau here. I would like to thank and acknowledge Louis for this help and for sharing his material. the format of the tutorial has been inspired from Mar Gonzalez Porta of Embl-EBI. I also want to acknowledge Joel Fillon, Louis Letrouneau (again), Francois Lefebvre, Maxime Caron and Guillaume Bourque for the help in building these pipelines and working with all the various datasets.