By Robert Eveleigh, MSc., Mathieu Bourgey, PhD., and Guillaume Bourque, PhD.
In this biohackathon module, you are working at a genomic center sequencing a patient's genome to guide decisions in medical treatment. In this scenario, the patient is in dire need of treatment so the identifation of clinically relevant mutations is time sensitive. Therefore, your challenge is to successfully analyze the given sample and identify the mutations in the sample as quickly as possible. The new Illumina sequencer, the Illumina SuperDuper, provides files containing all the reads (fastq or bam) and your goal is to generate the file with the mutations (vcf).
We will be working with one sample from the CEPH 1463 pedigree sequenced by the Genome in a Bottle (GIAB) consortium.
Due to time and resource constraints, we will provide reads from chromosome 22 only.
This work is licensed under a Creative Commons Attribution-ShareAlike 3.0 Unported License. This means that you are able to copy, share and modify the work, as long as the result is distributed under the same license.
The initial structure of your folders should look like this:
<ROOT>
|--raw_reads
`---gcat_set_025 # directory containing raw_reads. Directory contains both fastq and bam files as well as bed file.
|-- bin/ # bin directory. Contains bash scripts to execute standard pipeline, and benchmarking (F1 score generation).
|-- benchmarking/ # benchmarking results. Contains high quality variants from the GIAB
The team will be utilizing McGill's HPC cluster: Guillimin. Accounts have been setup and credientials to login will be provided by the instructors.
All access to the nodes listed above is permitted only via ssh. Applications such as Putty (for Windows) or on the command-line from within Linux or Mac can be used to make the ssh connection.
ssh -l <username>@<hostname>
For example:
ssh class81@aw-4r13-n01.calculquebec.ca
Once the team has access to the HPC, the team will need to setup the mugqic pipeline environment to allow the loading of modules for the programs you will be employing.
To setup enviroment please add the following lines to your ~/.bash_profile and then source your bash_profile to initiate changes. For details see Quick setup for abacus, guillimin and mammouth users
More information about genomic resources can be found here
All files required for this module have been uploaded to the shared space provide by McGill HPC
Commands:
##In home directory
cp -R /sb/project/class/BioHack/biohackathon_mutations/ ./
##Test execution script
cd biohackathon_mutations/
cp bin/c3g_execution_script.sh ./
time ./c3g_execution_script.sh
Commands to view all modules/programs available, load/unload a module and identify which modules currently loaded are as follows:
module avail
module load mugqic/GenomeAnalysisTK/3.7
module unload mugqic/GenomeAnalysisTK/3.7
module list
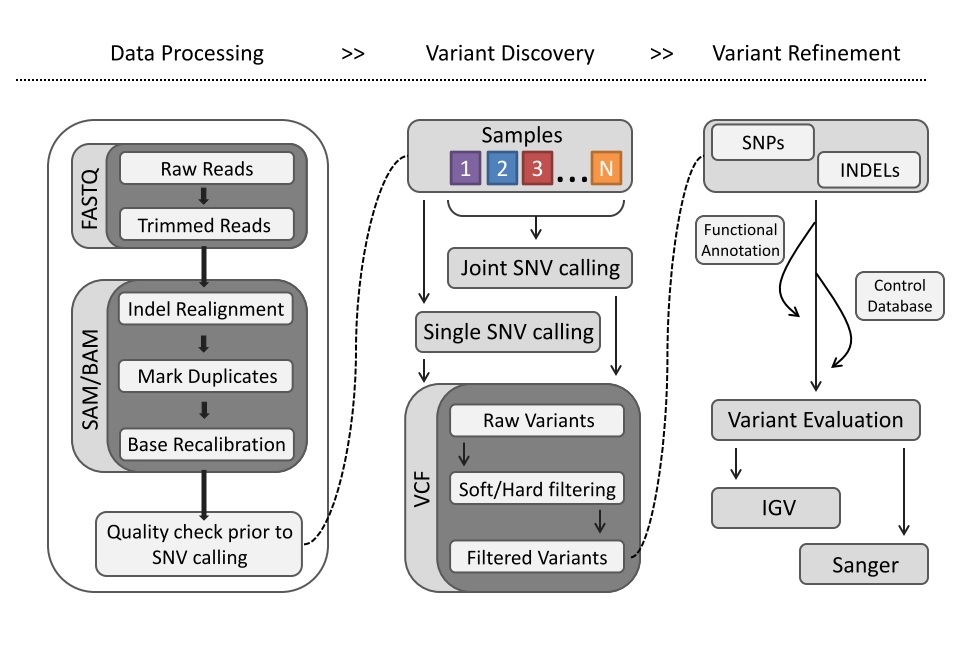
#Data processing and variant discovery
The sample from the CEPH pedigree has been sequenced. Given the raw data, C3G has processed the data using the GATK's 'best practices' as ground truth. This includes read trimming, quality reads mapped to the reference genome (GRCh37) and the resulting bam files have been further processed using indel realigner, fixmates, mark duplicates and base recalibration to reduce known systematic biases. Variants were called with GATK haplotype caller in GVCF mode.
The team's goal is to achieve similar F1 scores while decreasing computational time.
Processing time: Total CPU time ~13 mins (user + system time)
| True-pos | False-pos | False-neg | Precision | Sensitivity | F-measure |
|---|---|---|---|---|---|
| 553 | 36 | 167 | 0.9389 | 0.7681 | 0.8449 |
Teams will be ranked based on the number of false positive (FP), false negative (FN) and the (user + system) time required to generate the final vcf file.
- Have fun!
- The team may add/remove steps as well as swap in different programs at any step in the process. If the program you wish to add/try is not in our list of modules. Please install them in your local space.
- The team may start from the raw fastq files or the bam containing the raw reads. However, in the case of the latter, the team will be penalized the time required to align the reads (~75 seconds).
- Their will be a penalty of 10 seconds to cpu time for each FP and FN.
- A functioning bash script used to execute your commands.
- Execution time (user + system time) generated from the time unix command.
- Final vcf.
The following instructors will respond to emails between (9am - 10pm)
Robert Eveleigh: robert.eveleigh@mcgill.ca
Mathieu Bourgey: mathieu.bourgey@mcgill.ca
Guillaume Bourque: guil.bourque@mcgill.ca
We would like to acknowledge McGill HPC, Compute Canada and Calcul Quebec for their continued support and resources for this biohackathon module.