Uncertainty Quantification with Deep GLM Ensembles, including a PyTorch implementation of the CRISPRon architecture.
This notebook provides and implementation of Deep Ensembles (Lakshminarayanan et al., 2017) for the case of univariate regression, encompassing the case for Normal, Beta, and Gamma response variables for (-Inf, Inf), (0,1), and (0, Inf) data, respectively. The reported uncertainty are quantiles of the predictive distribution from the equally weighted ensemble. The latter is estimated numerically via simulating n_samples from the predictive distribution of each ensemble member (model), and then reporting the empirical quantiles of the pooled samples.
Included is a PyTorch implementation of the CRISPRon architecture for guideRNA design, as described in Xiang, Xi, et al. "Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning." Nature communications 12.1 (2021): 3238., which is a multimodal neural net architecture that takes as input a sequence and a static feature vector.
Accompanying the model is a custom DataSet class for the case where a single model input is (sequence, features), where sequence is a vector-valued sequence, and features is a vector of features, as is the case for CRISPRon.
Just run the cells that define the custom classes, and then use is reasonably simple.
myData = Seqs_and_Features(train_S, train_F, train_y)
myEnsemble = RegressionDeepEnsemble(BaseNet=CRISPRnet, dataset=myData,
n_estimators=10, batch_size=100,
response_var = torch.distributions.Beta)
myEnsemble.train_ensemble(n_epochs=20)
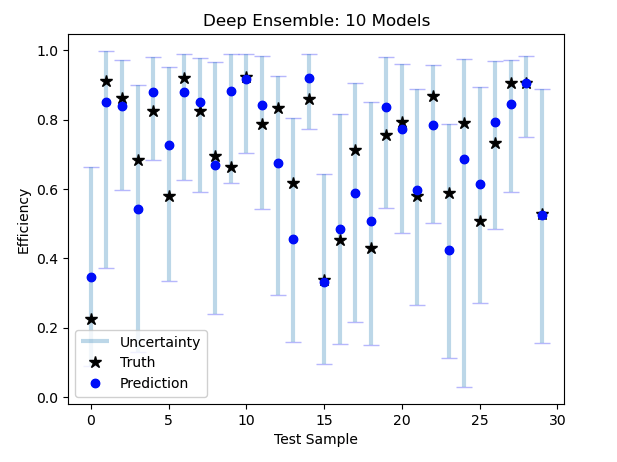
myEnsemble.predict(inputs = (test_S, test_F))There is also a built in tool for visualisation of uncertainties:
myEnsemble.plot_uncertainties(inputs=(S_test, F_test),
true_vals = y_test, plot_means=False,
lower=0.01, upper=0.99, n_samples=500)